The goal of biolabr is to provide a consistent location to store and access R functions that I have found to be useful for various things in the laboratory. The functions contained within this package will appear to be somewhat eclectic, but they will all be intended to provide a solution for various lab related activities that seem to reoccur and aren’t complex enough to require a dedicated package on their own.
You can install the up to date version of the package from github using devtools.
library(devtools)
install_github("baynec2/biolabr")This is a simple function that calculates R2 values for linear models split by a factor. I find this to be useful for displaying the R2 values for ggplots with multiple facets.
library(biolabr)
#> Loading required package: sangeranalyseR
#> Loading required package: stringr
#> Loading required package: ape
#> Warning: package 'ape' was built under R version 4.1.2
#> Loading required package: Biostrings
#> Loading required package: BiocGenerics
#>
#> Attaching package: 'BiocGenerics'
#> The following objects are masked from 'package:stats':
#>
#> IQR, mad, sd, var, xtabs
#> The following objects are masked from 'package:base':
#>
#> anyDuplicated, append, as.data.frame, basename, cbind, colnames,
#> dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep,
#> grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget,
#> order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
#> rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply,
#> union, unique, unsplit, which.max, which.min
#> Loading required package: S4Vectors
#> Warning: package 'S4Vectors' was built under R version 4.1.3
#> Loading required package: stats4
#>
#> Attaching package: 'S4Vectors'
#> The following objects are masked from 'package:base':
#>
#> expand.grid, I, unname
#> Loading required package: IRanges
#> Loading required package: XVector
#> Loading required package: GenomeInfoDb
#> Warning: package 'GenomeInfoDb' was built under R version 4.1.2
#>
#> Attaching package: 'Biostrings'
#> The following object is masked from 'package:ape':
#>
#> complement
#> The following object is masked from 'package:base':
#>
#> strsplit
#> Loading required package: DECIPHER
#> Loading required package: RSQLite
#> Loading required package: parallel
#> Loading required package: reshape2
#> Loading required package: phangorn
#> Warning: package 'phangorn' was built under R version 4.1.2
#> Loading required package: sangerseqR
#> Warning: package 'sangerseqR' was built under R version 4.1.3
#> Loading required package: gridExtra
#>
#> Attaching package: 'gridExtra'
#> The following object is masked from 'package:BiocGenerics':
#>
#> combine
#> Loading required package: shiny
#> Warning: package 'shiny' was built under R version 4.1.2
#> Loading required package: shinydashboard
#>
#> Attaching package: 'shinydashboard'
#> The following object is masked from 'package:graphics':
#>
#> box
#> Loading required package: shinyjs
#>
#> Attaching package: 'shinyjs'
#> The following object is masked from 'package:shiny':
#>
#> runExample
#> The following object is masked from 'package:RSQLite':
#>
#> show
#> The following object is masked from 'package:Biostrings':
#>
#> show
#> The following object is masked from 'package:GenomeInfoDb':
#>
#> show
#> The following object is masked from 'package:XVector':
#>
#> show
#> The following object is masked from 'package:IRanges':
#>
#> show
#> The following object is masked from 'package:S4Vectors':
#>
#> show
#> The following object is masked from 'package:stats4':
#>
#> show
#> The following objects are masked from 'package:methods':
#>
#> removeClass, show
#> Loading required package: data.table
#>
#> Attaching package: 'data.table'
#> The following objects are masked from 'package:reshape2':
#>
#> dcast, melt
#> The following object is masked from 'package:IRanges':
#>
#> shift
#> The following objects are masked from 'package:S4Vectors':
#>
#> first, second
#> Loading required package: plotly
#> Loading required package: ggplot2
#> Warning: package 'ggplot2' was built under R version 4.1.2
#>
#> Attaching package: 'plotly'
#> The following object is masked from 'package:ggplot2':
#>
#> last_plot
#> The following object is masked from 'package:XVector':
#>
#> slice
#> The following object is masked from 'package:IRanges':
#>
#> slice
#> The following object is masked from 'package:S4Vectors':
#>
#> rename
#> The following object is masked from 'package:stats':
#>
#> filter
#> The following object is masked from 'package:graphics':
#>
#> layout
#> Loading required package: DT
#> Warning: package 'DT' was built under R version 4.1.2
#>
#> Attaching package: 'DT'
#> The following objects are masked from 'package:shiny':
#>
#> dataTableOutput, renderDataTable
#> Loading required package: zeallot
#> Loading required package: excelR
#> Loading required package: shinycssloaders
#> Loading required package: ggdendro
#> Warning: package 'ggdendro' was built under R version 4.1.2
#> Loading required package: shinyWidgets
#> Warning: package 'shinyWidgets' was built under R version 4.1.2
#>
#> Attaching package: 'shinyWidgets'
#> The following object is masked from 'package:shinyjs':
#>
#> alert
#> Loading required package: openxlsx
#> Loading required package: tools
#> Loading required package: rmarkdown
#> Warning: package 'rmarkdown' was built under R version 4.1.2
#> Loading required package: knitr
#> Warning: package 'knitr' was built under R version 4.1.2
#> Loading required package: seqinr
#> Warning: package 'seqinr' was built under R version 4.1.2
#>
#> Attaching package: 'seqinr'
#> The following object is masked from 'package:shiny':
#>
#> a
#> The following object is masked from 'package:sangerseqR':
#>
#> read.abif
#> The following object is masked from 'package:Biostrings':
#>
#> translate
#> The following objects are masked from 'package:ape':
#>
#> as.alignment, consensus
#> Loading required package: BiocStyle
#>
#> Attaching package: 'BiocStyle'
#> The following objects are masked from 'package:rmarkdown':
#>
#> html_document, md_document, pdf_document
#> The following object is masked from 'package:shiny':
#>
#> markdown
#> Loading required package: logger
#> Welcome to sangeranalyseR
#> Loading required package: rBLAST
library(ggplot2)
data(mtcars)
R2 = facet_R2(mtcars,y = "mpg",x = "hp",by = "gear")
p1 = ggplot(mtcars,aes(mpg,hp))+
geom_point()+
geom_smooth(method = "lm")+
facet_grid(~gear)+
geom_text(data = R2,aes(x = 30, y = 300,label = R2))
p1
#> `geom_smooth()` using formula 'y ~ x'This function allows the user to parse CFU spot plating data that is recorded in an excel sheet (found in /template_files/format_CFU/). There are two types of templates, one for serial dilutions that are made across plates and another that is used for serial dilutions that are made row wise.
If you need to add plates to the template, just copy and paste more copies of the cells representing a plate directly below the previous ones.
library(biolabr)
t1 = format_CFU("example_files/CFU_Dilutions_On_Different_Rows_Example.xlsx")
#> New names:
#> New names:
#> • `` -> `...1`
#> • `` -> `...2`
#> • `` -> `...3`
#> • `` -> `...4`
#> • `` -> `...5`
#> • `` -> `...6`
#> • `` -> `...7`
#> • `` -> `...8`
#> • `` -> `...9`
#> • `` -> `...10`
#> • `` -> `...11`
#> • `` -> `...12`
#> • `` -> `...13`
#> • `` -> `...14`
#> • `` -> `...15`
#> • `` -> `...16`
#> • `` -> `...17`
#> • `` -> `...18`
head(t1)
#> Sample_ID PlateNumber Dilution Media Condition Length_of_Incubation Well
#> 1 TEST.A1 TEST 3 MTGE Aerobic 24 A1
#> 2 TEST.A2 TEST 3 MTGE Aerobic 24 A2
#> 3 TEST.A3 TEST 3 MTGE Aerobic 24 A3
#> 4 TEST.A4 TEST 2 MTGE Aerobic 24 A4
#> 5 TEST.A5 TEST 2 MTGE Aerobic 24 A5
#> 6 TEST.A6 TEST 2 MTGE Aerobic 24 A6
#> Colony_Count Dilution_Factor CFU_mL
#> 1 12 1000 6000000
#> 2 11 1000 5500000
#> 3 10 1000 5000000
#> 4 4 100 200000
#> 5 3 100 150000
#> 6 3 100 150000This function is useful for programmatically generating consensus sequences from Genewiz 16S results (https://www.genewiz.com/en/Public/Services/Molecular-Genetics/Cell-Line-Authentication) and BLASTing them against a database to determine the taxonomy of your sample.
Unfortunately, this requires the user to have the BLAST+ command line tools downloaded on their system. Even more unfortunately, this also requires download of a large sql database (~26 Gb) that is used to map the NCBI accession numbers to the actual taxonomy.
Still, I find this beats manually copying and pasting sequences into BLAST.
First, you need to install BLAST+ on your system. This can be accomplished by following the instructions below (For Mac OS).
-
Click the link to download the appropriate installer from https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/
-
Double click ncbi-blast-2.4.0+.pkg. If you see the dialog below, Hold down the Control key and click ncbi-blast-2.4.0+.pkg. From the contextual menu choose Open.
-
By default the BLAST+ applications will be installed in /usr/local/ncbi/blast, overwriting its previous contents (an uninstaller is provided and it is recommended when upgrading a BLAST+ installation).
Next, you will need to download the 16SMicrobial database
- This can be found by going to ftp://ftp.ncbi.nlm.nih.gov/blast/db/v4/
- I like to store this in the same location as the blast files (/usr/local/ncbi/blast/db)
Lastly, you will need to install the sql database used to convert the NCBI_IDs to taxonomy. This can be done by using the taxonomizer R package
## Run this command in the directory that you want to store the database in
setwd("Location you want to download accession database to")
taxomomizer::prepareDatabase('accessionTaxa.sql')Once all of these steps have been completed you should be able to run the function on your data.
Importantly, this function will return the lowest level of taxonomy that is completely unambiguous at your specified percent identity. This means if you specify a percent identity of 99%, and there are matches to Escherichia coli and Escherichia albertii, the function will only identify your taxa at the genus level (Escherichia).
#Note that the file paths specified here are for my machine. Depending on how you set yours up the file paths will differ.
library(biolabr)
taxonomy = Genewiz_16S(folder_path = "example_files/Genewiz_16S_Data/",
blast_db_path = "/usr/local/ncbi/blast/db/16SMicrobial/16SMicrobial",
accessionToTaxa_path ="/Volumes/kaleidobio/Shared/D2/Departments/Research/Biology/biolabr/accessionTaxa.sql",
similarity = 99)
DT::datatable(taxonomy)Here we can see that all of these isolates named CB011 - CB021 could not be unambiguously identified beyond order.
This function is useful for parsing data from the Ankom Gas System and getting it into a tidy format. (https://www.ankom.com/product-catalog/ankom-rf-gas-production-system).
The original output files are in multiple sheets of an excel workbook, this function consolidates all of this information to a tidy data frame.
library(biolabr)
df = parse_gas_data("example_files/Ankom_Gas_Data.xls")
head(df)
#> # A tibble: 6 × 6
#> Time_hr Sample.ID Cumulative_Pressure Absolute_Pressure Temperature Battery_…¹
#> <dbl> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 0 Reference 14.5 14.5 34.8 6.95
#> 2 0 Sample1 0 14.4 33.2 6.87
#> 3 0 Sample2 0 14.4 32.3 6.93
#> 4 0 Sample3 0 14.4 32.3 6.90
#> 5 0 Sample4 0 14.4 32.4 6.90
#> 6 0 Sample5 0 14.4 32.0 6.93
#> # … with abbreviated variable name ¹Battery_VoltageThis function simpily removes leading 0s from Well IDS. For example, this will change A01 to A1. Some machines in the lab export well IDs with leading 0s, others do not. As such, this function is frequently useful for mapping two data sources that have Well IDs in differing formats.
library(biolabr)
machine_data_leading_0 = read.csv("example_files/Well_IDs_with_leading_0.csv")
head(machine_data_leading_0,2)
#> Well Fluor End.RFU
#> 1 A01 SYBR 5938.963
#> 2 A02 SYBR 6665.284
machine_data_no_leading_0 = machine_data_leading_0 %>%
dplyr::mutate(Well = rm_leading_0(Well))
head(machine_data_no_leading_0,2)
#> Well Fluor End.RFU
#> 1 A1 SYBR 5938.963
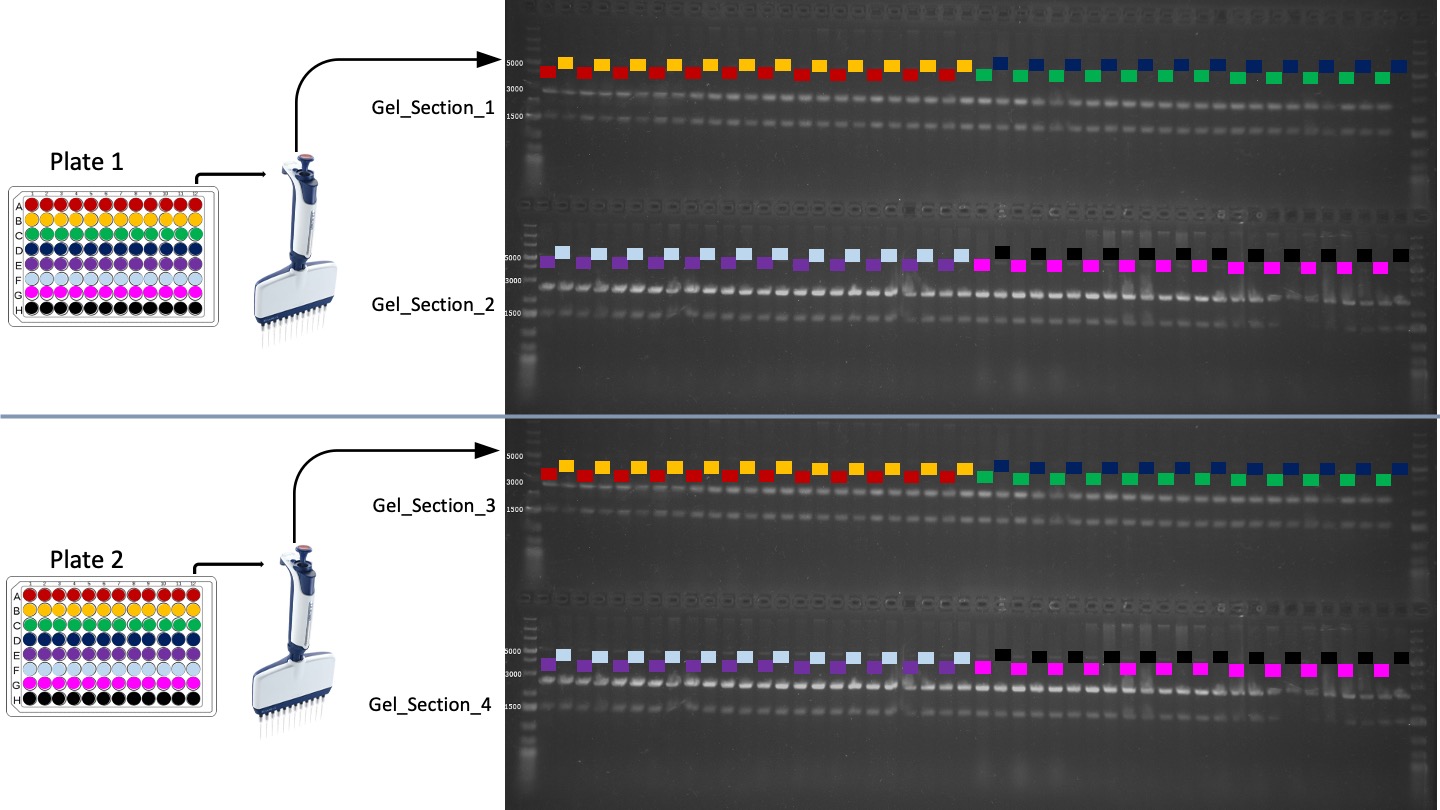
#> 2 A2 SYBR 6665.284This function is useful for assigning Well IDs to DNA bands that have been run through gel electrophoresis, and then detected using the open source GelAnalyzer software (http://www.gelanalyzer.com/).
This function assumes that samples have been loaded onto the gel using a multichannel pipette following a specific pattern. One single large gel can accommodate up to 2 x 96 well plates. This is displayed visually below:
d1 = assign_gel_wells("example_files/Gel_Band_Data.xlsx")
head(d1)
#> # A tibble: 6 × 6
#> Well_ID Gel_Section Band_MW Rf `Raw volume` intensity
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 E1 2 5147 0.264 551 145.
#> 2 E1 2 2573 0.425 1761 748.
#> 3 E1 2 1537 0.559 1243 695.
#> 4 F1 2 5059 0.268 626 168.
#> 5 F1 2 2494 0.433 2042 884.
#> 6 F1 2 1516 0.563 1486 837.This is an operator for negated value matching. I frequently find it useful to have an operator that is the opposite of %in%. This operator returns FALSE if a match is present, TRUE if a match is not present.
test = c(1,1,1,1,1,2,1,1,1,1)
test %nin% 2
#> [1] TRUE TRUE TRUE TRUE TRUE FALSE TRUE TRUE TRUE TRUEThis package also contains functions designed to make it easy to do dynamic time warping in order to look through high dimensional datasets for features that match a given pattern of interest.
Here in this example we will be looking at an untargeted metabalomic data set of a timeseries of unique IDs described as a m/z at a given retention time. The time series includes a measurement at 0, 4.00, 8.03, , and 39.44 hrs.
First let’s generate a hypothetical pattern of interest and bind it to our data set
# Pattern of interest starts out high then rapidly decreases after ~4 hrs.
POI = data.frame(POI = c(100000,80000,1000,0,0,0))
mb = readr::read_csv("example_files/Metabolomic_Time_Series.csv")
#> Rows: 6 Columns: 1515
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> dbl (1515): M165T356, M234T171, M177T411, M468T203, M380T447, M313T199, M518...
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
all = cbind(mb,POI)Now let’s apply the dynamic time warping algorithm to generate a distance matrix.
#Note that this function scales the data within each time series by default.
dist = biolabr::dtwarper(all)
saveRDS(dist,"example_files/dist.rds")#reading from file to save time.
dist = readRDS("example_files/dist.rds")Note that this function takes a while to run as dynamic time warping is somewhat computationally intense.
Now we can pull out the 10 features that match this pattern the best.
n_IDs = find_n_similar(dist,"POI", n = 10)Let’s plot these IDs so we can see what the trends look like.
library(ggplot2)
# First need to convert the format back to long so we can plot these easily
Time = tibble::tibble(Time = c(0,4,8.03,22.43,25.43,29.43))
# Filtering to keep the 10 closest matches
long = cbind(Time,all) %>%
tidyr::pivot_longer(2:length(.)) %>%
dplyr::filter(name %in% c(n_IDs,"POI"))
# Plotting
p1 = long %>%
ggplot(aes(Time,value,color = name))+
geom_point()+
geom_line()+
facet_wrap(~name,scales = "free_y")+
theme(legend.position = "none")+
ggtitle("10 Most Similar Features to POI by Dynamic Time Warping")
p1This package also contains a simple template that can be used for RMD analysis files called Kaleido Analysis.