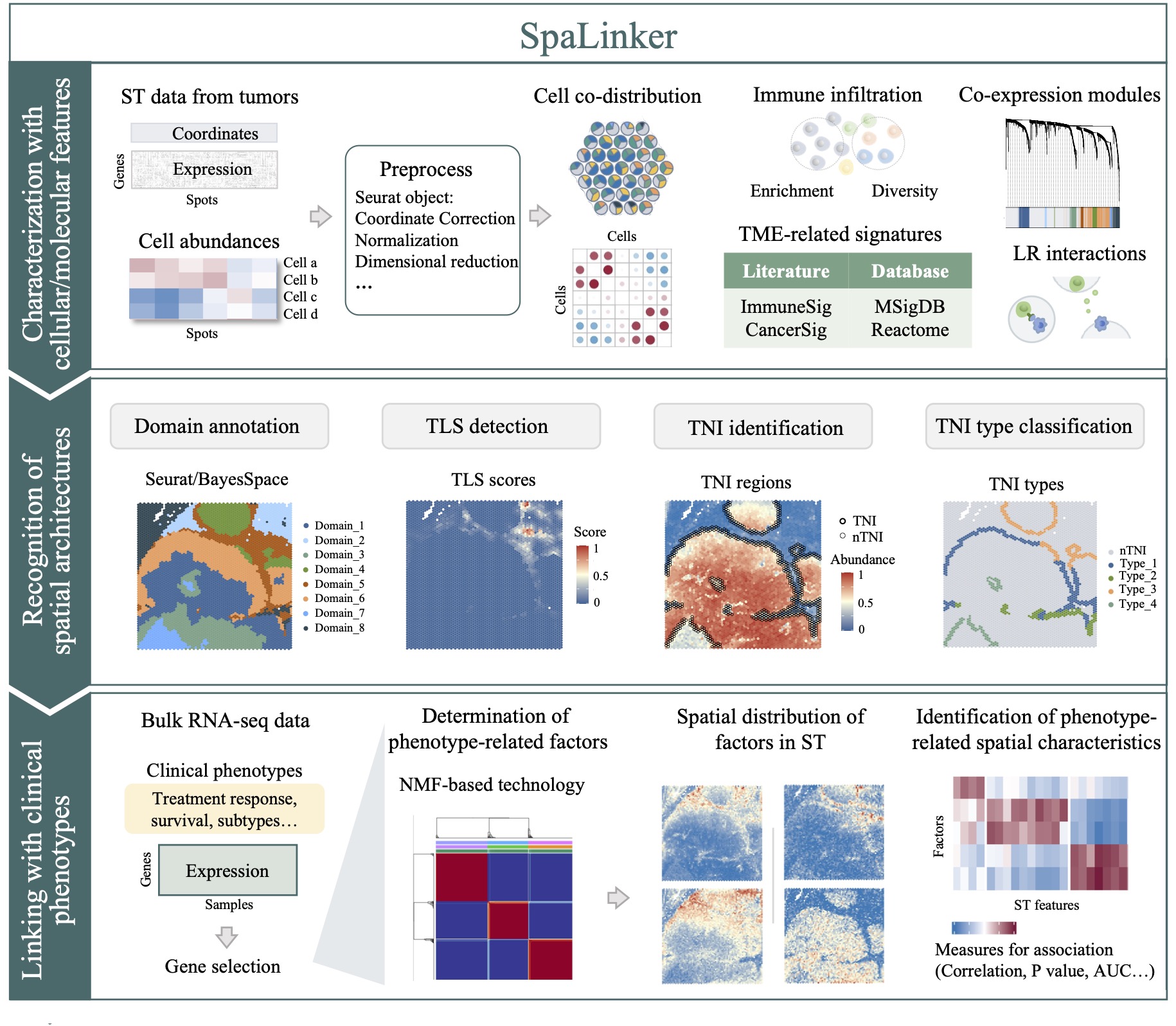
 SpaLinker is an integrative analytical framework designed for TME deciphering and phenotype linking using spot-resolution ST data. It consists of three functional parts (Overview): (1) Characterization of the TME with molecular and cellular features; (2) Recognition of spatial architectures; and (3) Linking with clinical phenotypes.
SpaLinker is an integrative analytical framework designed for TME deciphering and phenotype linking using spot-resolution ST data. It consists of three functional parts (Overview): (1) Characterization of the TME with molecular and cellular features; (2) Recognition of spatial architectures; and (3) Linking with clinical phenotypes.
SpaLinker has been built and tested with R >= 4.3.2. Specific package dependencies are defined in the package DESCRIPTION.
# Install devtools, if necessary
if (!requireNamespace("devtools", quietly = TRUE))
install.packages("devtools")
devtools::install_github("bm2-lab/SpaLinker")
For examples of typical SpaLinker usage, please see our package vignette and download the test data.