Challenge: CORD-19
Task: What do we know about COVID-19 risk factors?
Submission: Kaggle notebook
- Adrianna Safaryn
- Anna Haratym-Rojek
- Cezary Szulc
- Marek Grzenkowicz
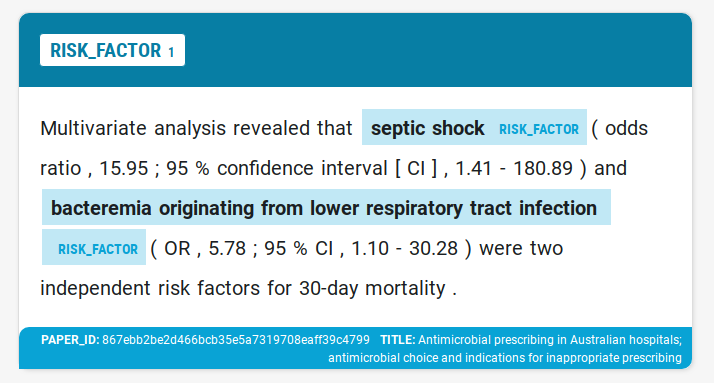
We wanted to use named entity recognition (NER) to highlight names of risk factors (RF). Our goal was training a custom NER model for spaCy, that could later be use to recognize risk factors in medical publications.
- Data preprocessing to extract 'risk factor(s)' sentences
- Manual data annotation in Prodigy
- Pretraining different models - we experimented with different base models and trained a number of
tok2vec layers to maximize the F-score
- base models:
en_vectors_web_lg,en_core_web_lg,en_core_sci_lg - tok2vec layers were trained for: RF sentences, subset of abstracts, all abstracts
- base models:
- Labelling more data by correcting the predictions of the top model trained in the previous step
- Go back to step #3 to pretrain a new model using more data and then label even more data
- Training the final model with all gathered annotations
Each iteration uses all datasets from the previous one and adds more annotations. For detailed information about every trained model, see the notebook train_experiments_2.ipynb.
| Iteration | Datasets (data/annotated/) | Best F-score |
|---|---|---|
| 1 | cord_19_rf_sentences |
53.333 |
| 2 | above + cord_19_rf_sentences_correct |
75.630 |
| 3 | above + cord_19_rf_sentences_correct_2 |
74.894 |
| 4 | above + cord_19_rf_sentences_correct_3 |
68.770 |
| Iteration | Datasets (data/annotated/) | Best F-score | Download |
|---|---|---|---|
| 1 | cord_19_rf_sentences |
57.778 | en_ner_rf_i1_md |
| 2 | above + cord_19_rf_sentences_correct |
74.380 | en_ner_rf_i2_md |
| 3 | above + cord_19_rf_sentences_correct_2 |
74.236 | en_ner_rf_i3_md |
| 4 | above + cord_19_rf_sentences_correct_3 |
69.725 | en_ner_rf_i4_md |
Using a smaller base model (md instead of lg) results in significantly smaller model, while the F-score
moves in both directions depending on the iteration.
Medium models for iterations 1..4 can be installed using the download links from the table above.
The directory test/ contains a demo of the models in action (separate Conda environment + notebook).
- Training of tok2vec layers: Kaggle notebook
- Full set of annotations:
- cord_19_rf_sentences_merged.jsonl (dump of the Prodigy dataset)
- cord_19_rf_sentences_merged.json (spaCy JSON format)
- Log of all experiments (including data annotation and model training): train_experiments_2.ipynb
- Early experiments: train_experiments_1.ipynb
- Detailed discussion posted at the Kaggle forum: Custom NER model to recognize risk factor names
- Question posted to the Prodigy support forum: Annotating compound entity phrases
+------------------+ +------------------+ +------------------+
| | | | | | +----------------------------+
| | FILTER | Abstracts and | FILTER | Sentences that | | |
| CORD-19 dataset +---------->| articles that +---------->| contain phrase +--------> | cord_19_rf_sentences.jsonl |
| | | mention COVID-19 | | "risk factor(s) | | |
| | | and synonyms | | | +----------------------------+
+------------------+ +------------------+ +------------------+- Prodigy - text annotation
- spaCy - NLP and model training
- scispaCy - specialized spaCy models for biomedical text processing
- Miniconda - environment setup (you can use
conda env create -f environment.ymlto set up the Python environment with all packages and models)
COVID-19 Open Research Dataset (CORD-19). 2020. Version 2020-03-13.
Retrieved from https://pages.semanticscholar.org/coronavirus-research.
Accessed 2020-03-26. doi:10.5281/zenodo.3715506