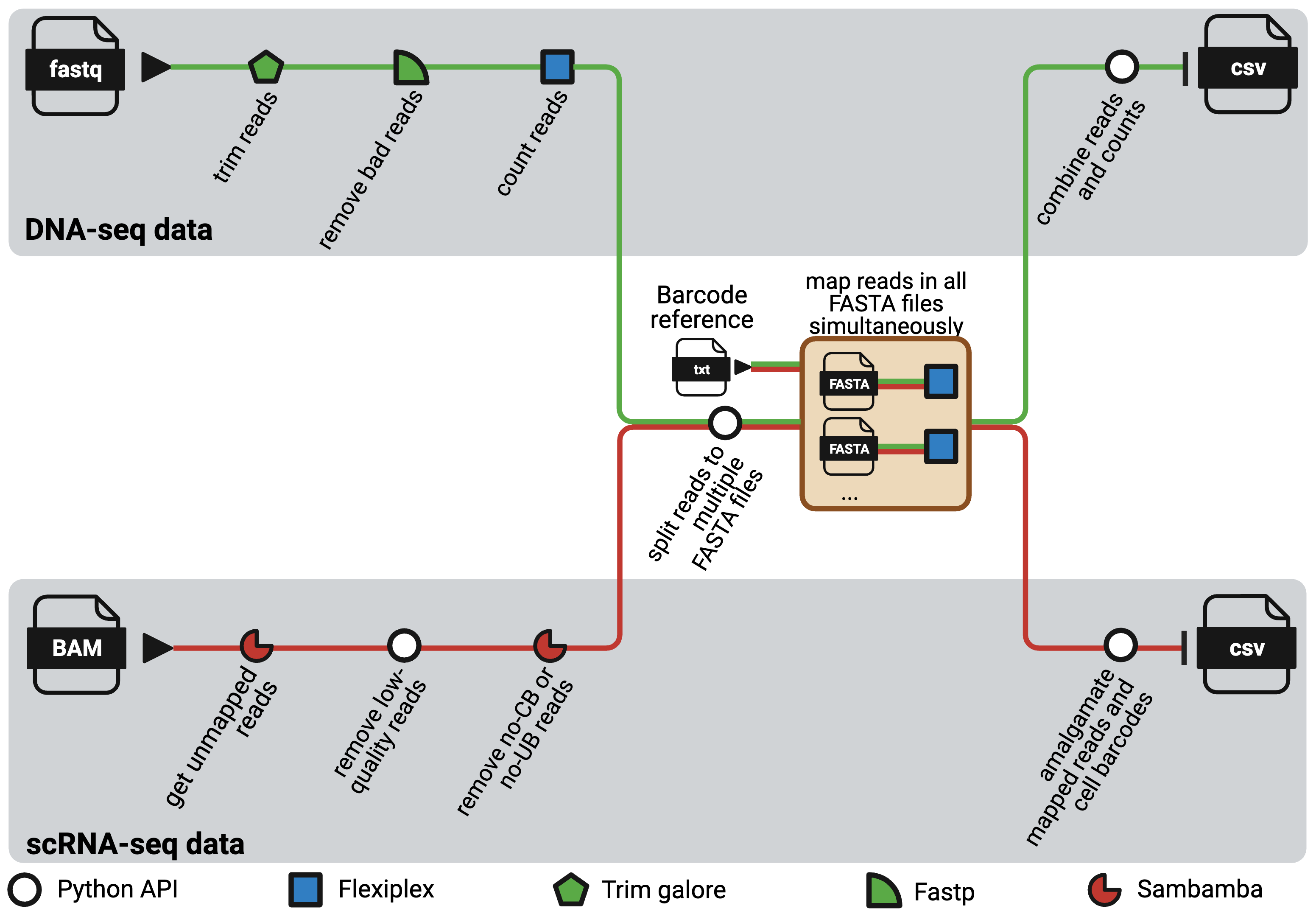
NextClone is a Nextflow pipeline to facilitate rapid extraction and quantification of clonal barcodes from both DNA-seq and scRNAseq data. DNA-seq data refers to dedicated DNA barcoding data which exclusively sequences the synthetic lineage tracing clone barcode reads using Next Generation Sequencing.
The pipeline comprises two distinct workflows, one for DNA-seq data and the other for scRNAseq data. Both workflows are highly modular and adaptable, with software that can easily be substituted as required, and with parameters that can be tailored through the nextflow.config file to suit diverse needs. It is heavily optimised for usage in high-performance computing (HPC) platforms.
git clone this repo
Make a conda/python venv and install biopython, pysam, pandas, and numpy
Have Flexiplex, cutadapt, FastQC, fastp, Trim galore, and sambamba in PATH
To run on 10X STICR data the alignment/cellranger for the STICR and transcriptome library needs to be done together!
If you're using PIP-seq make sure to realign the trimmed barcoded fastqs from PIPSeeker with STARSolo with unmapped reads within the bam
STARSolo example run inside a PIPSeeker output folder:
# Assuming out is the pipseeker output folder
R1=$(R1=$(ls barcoded_fastqs/*R1*); echo $R1 | sed 's/ /,/g');R2=$(R2=$(ls barcoded_fastqs/*R2*); echo $R2 | sed 's/ /,/g')
STAR --genomeDir ~/human_GRCh38_optimized_reference_v2_STAR --runThreadN 16 --soloType CB_UMI_Simple --soloCBstart 1 --soloCBlen 16 --soloUMIstart 17 --outSAMattributes CB CR CY GX GN UB UR UY NH HI nM AS sF --outSAMtype BAM SortedByCoordinate --soloCBmatchWLtype Exact --soloUMIdedup 1MM_CR --soloFeatures Gene SJ GeneFull GeneFull_Ex50pAS GeneFull_ExonOverIntron Velocyto --soloMultiMappers EM --soloCellReadStats Standard --soloCellFilter EmptyDrops_CR --soloUMIfiltering MultiGeneUMI_CR --outSAMunmapped Within --soloBarcodeReadLength 0 --readFilesCommand zcat --limitBAMsortRAM 1775716961230000 --soloCBwhitelist barcodes/barcode_whitelist.txt --soloUMIlen 12 --readFilesIn $R2 $R1 --outFileNamePrefix trimmed_
You need the STICR whitelist in a specific format that has all 3 possible indices and truncated down to 58 bps (minimum length for bit 3 demux)
For convenience I've uploaded to google drive gzipped whitelist, you will need to unzip it. The file is ~21 GB unzipped.
Link: https://drive.google.com/file/d/1FqhcDpYlQ1qbT5pK__skXZ3N3QCrm1-T/view?usp=sharing
Modify the nextflow.config file for the STICR whitelist path (clone_barcodes_reference) and/or output folder run which is set to current dir where you run the nextflow run command:
Depending on the compute availability, modify the regular_mapping parameters for memory and CPU usage.
Run once in the bam/cellranger out folder (here it assumes NextClone is in home dir):
nextflow run ~/NextClone/main.nf -c ~/NextClone/nextflow.config