DigiCam pipeline based on ctapipe
The documentation can be found here: Digicampipe documentation
- install Anaconda
git clone https://github.com/cta-sst-1m/digicampipeconda env create -f digicampipe/environment.ymlsource activate digicampipepip install -e digicampipepytest digicampipe/digicampipe
You'll need Anaconda, so if you don't have it yet, just install it now. Follow the anaconda installation instructions. We propose to use the most recent version.
wget https://repo.continuum.io/archive/Anaconda3-5.0.0.1-Linux-x86_64.sh
bash ./Anaconda3-5.0.0.1-Linux-x86_64.sh
We propose to have a tidy place and clone digicampipe into a folder ctasoft/
mkdir ctasoft
cd ctasoft
git clone https://github.com/cta-sst-1m/digicampipe
To not mix up your anaconda root environment with digicampipe, we propose to make a so called environment, with all the dependencies in it.
conda env create -f digicampipe/environment.yml
This is usefull when new packages are added to the environment.yml
conda env update -f digicampipe/environment.yml
source activate digicampipe
Please Note: When working with digicampipe, please always make sure you are really using the digicampipe environment. After source activate digicampipe
your prompt should look similar to this this:
(digicampipe) username@host:~/ctasoft$
pip install -e digicampipe
Note for MacOS users at the moment the installation of the protozfitsreader package requires some manual work.
Please type this command before running pip
export DYLD_LIBRARY_PATH=$DYLD_LIBRARY_PATH:<PATH_TO_YOUR_ANANCODA_LIBRARY_PATH>/lib/python3.6/site-packages
<PATH_TO_YOUR_ANANCODA_LIBRARY_PATH> is where your anaconda was installed.
pytest digicampipe/digicampipe
Some tests depend on astrometry.net (see below) and take long, so by default you might want to skip them. If you want to execute all tests, please do:
pytest -c all_tests.ini
To write tests please follow the instructions in the Pytest-doc. It is important that you add the test resources to the repo. To do this add the resource path to "package_data=" in the setup.py file.
Build the documentation with Sphinx (optional)
In the digicampipe directory run:
cd docs/
make html
This should create the documentation in digicampipe/docs/build/html.
You can open the html files with your favorite web browser.
To delete the documentation us:
make clean
You may want to mount the data from a remote onto your local machine. To do this
you can use sshfs. Information about sshfs can be found here
sshfs
Usage example :
sudo sshfs -o allow_other <username>@<remote_adress>:<remote_path> <local_path>
With digicampipe comes the example program digicam-view. To use it, just go call it like this:
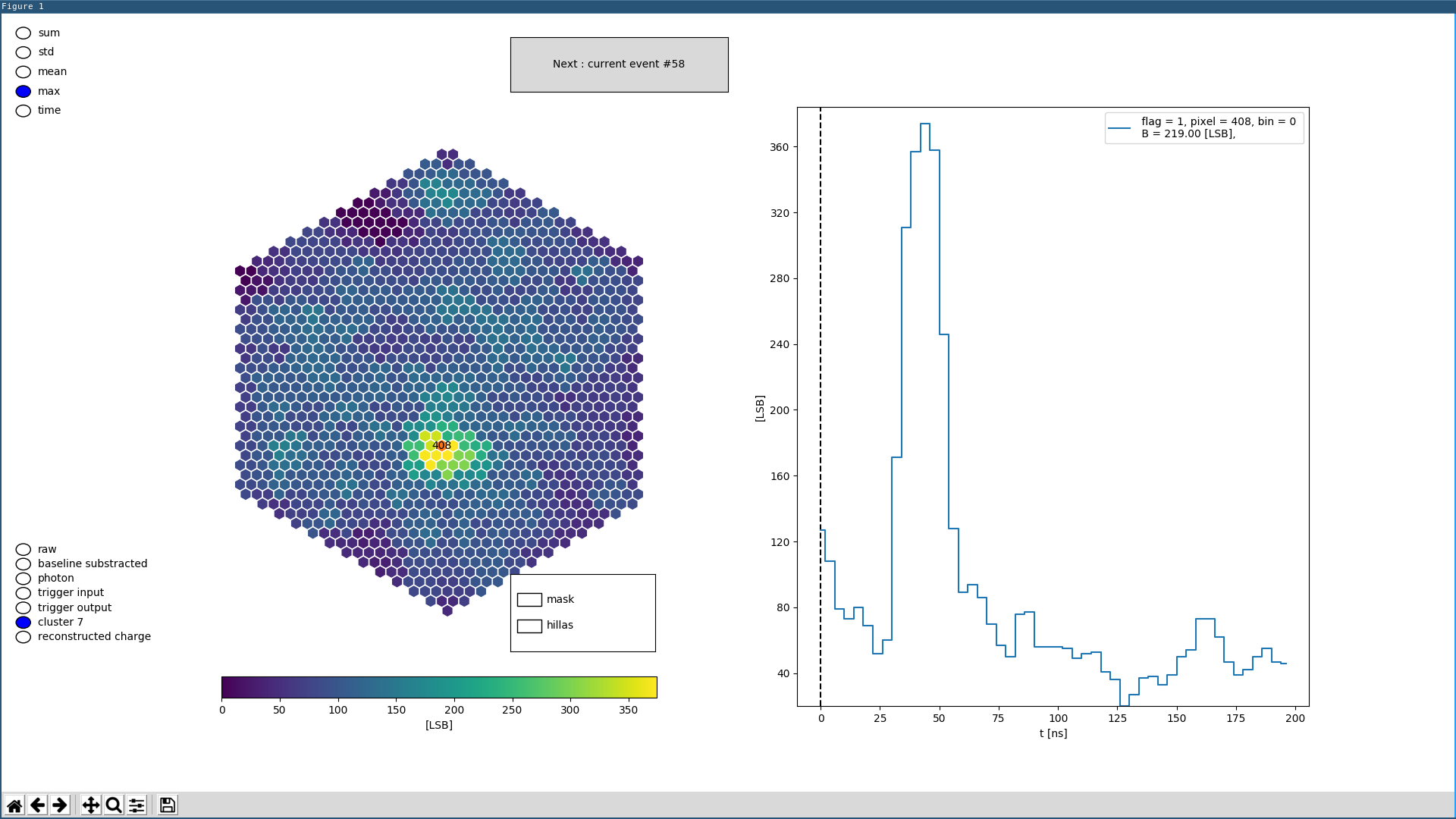
dneise@lair:~/sst/data$ digicam-view SST1M01_20171030.066.fits.fz
And you might see something like this:
To be able to determine the pointing using stars reflections on the lid, astrometry.net is needed. If not installed locally, the web service will be used instead. To install astrometry.net:
sudo apt-get install astrometry.net
You will need to download catalogs too:
mkdir ~/astrometry-data
cd ~/astrometry-data
wget http://broiler.astrometry.net/~dstn/4200/wget.sh
chmod +x wget.sh
./wget.sh
Grab a beer as it will take a while ... Also, indexes 4203 and lower are probably not needed.
Then add the folowing line to /etc/astrmetry.cfg:
add_path ~/astrometry-data
That's it, you are ready to go.
The purpose of the template scan analysis is to find the shape of the typical non-saturating pulse.
To perform this analysis download the needed raw data files from somewhere and put
them into someplace you like, then modify template_scan_analysis.sh so it finds the files.
Afterwards call:
./template_scan_analysis.sh
... and wait. This analysis needs 3 cores, ~6GB memory and takes on my machine ~25minutes. Once the analysis is done, you can look at the results and perform the higher level anlysis with:
jupyter-notebook template_scan_analysis.ipynb