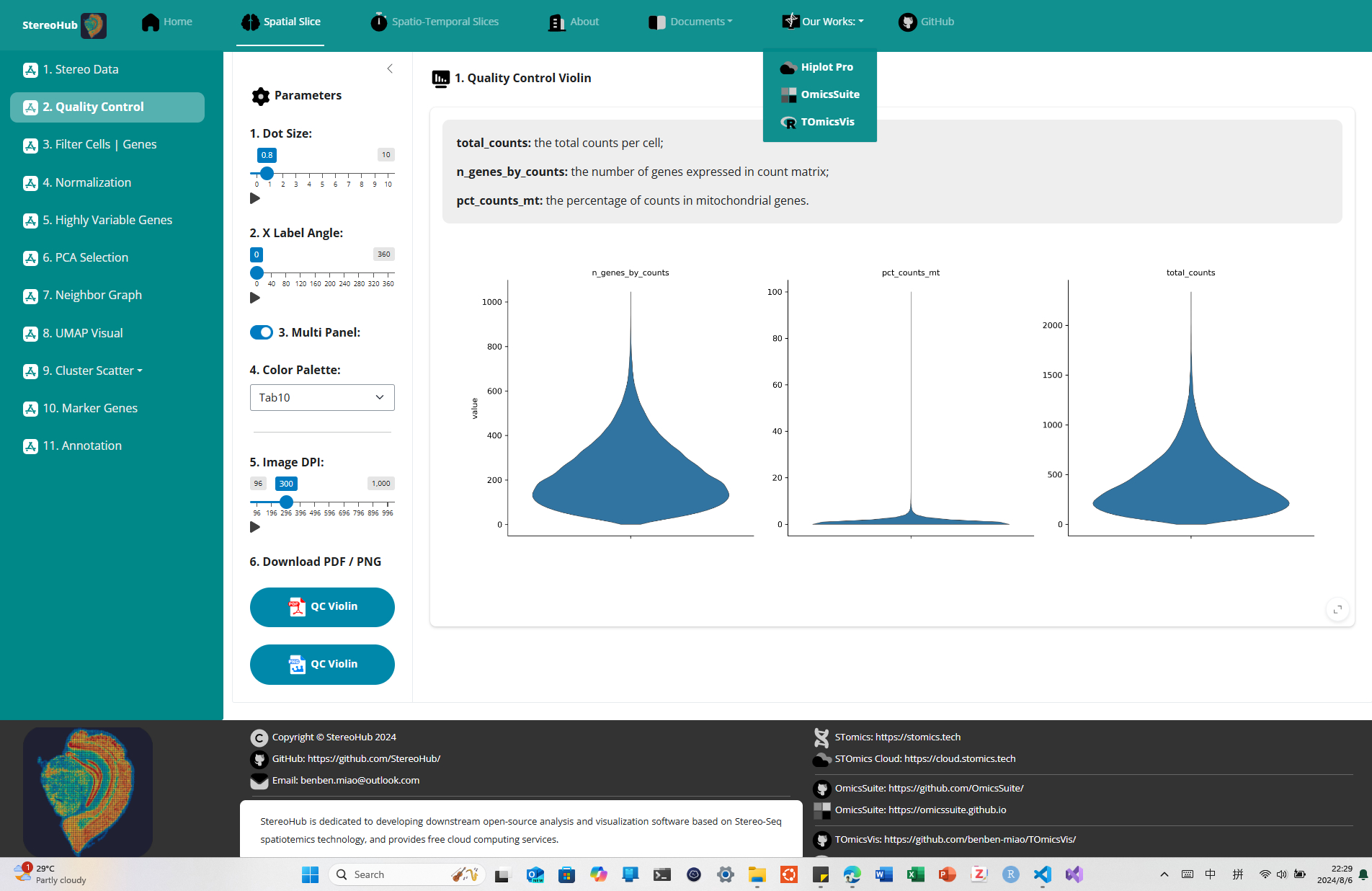
StereoHub: An interactive cloud platform for downstream analytics at Stereo-Seq. The StereoHub user interface is designed and developed based on the Shiny v1.0.0 framework of Python v3.8.10, providing rich web components for data manipulation, function parameters, and result display, making data analytics parameters as completely intuitive as possible for scientists. The functions of StereoHub for Stereo-Seq spatiotemics data analytics are realized by important functional modules such as stereopy v3.1.3, anndata v0.9.2, biopython v1.83.0, hdf5 v1.14.0, panel v0.14.4, bokeh v2.4.3, plotly v5.23.0, etc. Thanks to these excellent open-source modules.
a. Github Team: https://github.com/StereoHub/
b. Source Repository: https://github.com/StereoHub/StereoHub
c. Documents: https://stereohub.github.io
d. Cloud Platform: https://hiplot.com.cn/stereohub
a. STomics Stereo-Seq: https://stomics.tech
b. STomics Cloud: https://cloud.stomics.tech
c. STOmics Database: https://db.cngb.org/stomics/
d. ImageStudio, StereoMap: https://stomics.tech/products/BioinfoTools/OfflineSoftware
e. STomics Github: https://github.com/STOmics/
f. Stereopy: https://github.com/STOmics/Stereopy/
StereoHub Cloud: https://hiplot.com.cn/stereohub/
# 1. git clone repository
git clone https://github.com/StereoHub/StereoHub.git
cd StereoHub
# 2. Install Environment
# 2.1 For Windows
.\env-win.bat
# 2.2 For Linux
bash env-linux.sh
# 2.3 Install Steps
# 2.3.1 Mamba Env Create and Activate
mamba create -n stereohub python=3.8
mamba activate stereohub
# 2.3.2 Shiny
mamba install -c conda-forge shiny=1.0.0 shinywidgets=0.3.2 IPython=8.12.2 ipywidgets=8.1.3
# 2.3.3 Utils
mamba install -c conda-forge numpy=1.23.5 pandas=1.5.3 matplotlib=3.7.1 faicons=0.2.2
# 2.3.4 Stereopy
mamba install stereopy=1.3.1 -c stereopy -c grst -c numba -c conda-forge -c bioconda -c fastai -c defaults
# 2.4 Conda Env Export and Create
conda env create -f stereohub.yml
conda activate stereohub# 1. For Windows
.\start-win.bat
# 2. For Linux
bash start-linux.sh
# 3. All Terminals
python -m shiny run \
--host 127.0.0.1 \
--port 5000 \
--reload \
--reload-includes "*.py,*.css,*.js,*.html,*.md" \
--reload-excludes "*.png,*.pdf" \
--log-level info \
--app-dir "." \
--launch-browser \
--dev-mode \
app.pyShinylive: Shiny + WebAssembly: https://shiny.posit.co/py/docs/shinylive.html
shiny create myapp
pip install shinylive
shinylive export myapp site
python3 -m http.server --directory site 5000Self-hosted deployments: https://shiny.posit.co/py/docs/deploy-on-prem.html
# 1. shiny-server
wget https://download3.rstudio.org/centos7/x86_64/shiny-server-1.5.22.1017-x86_64.rpm
sudo rpm -ivh shiny-server-1.5.22.1017-x86_64.rpm
# 2. stereohub environment
bash env-linux.sh
# 3. shiny-server home
cd /srv/shiny-server/
git clone https://github.com/StereoHub/StereoHub.git
# 4. shiny-server config
sudo vim /etc/shiny-server/shiny-server.conf
python /cluster/envs/miniconda3/envs/stereohub/bin/python;
run_as shiny;
server {
listen 3838;
# Define a location at the base URL
location / {
# Host the directory of Shiny Apps stored in this directory
site_dir /srv/shiny-server;
# Log all Shiny output to files in this directory
log_dir /var/log/shiny-server;
# An index of the applications available in this directory will be shown.
directory_index on;
}
}
# 5. shiny-server manage
sudo systemctl start shiny-server
sudo systemctl status shiny-server
sudo systemctl restart shiny-serverAo Chen, Sha Liao, Mengnan Cheng, Longqi Liu, Xun Xu, Jian Wang. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell, 2022, doi: https://doi.org/10.1016/j.cell.2022.04.003