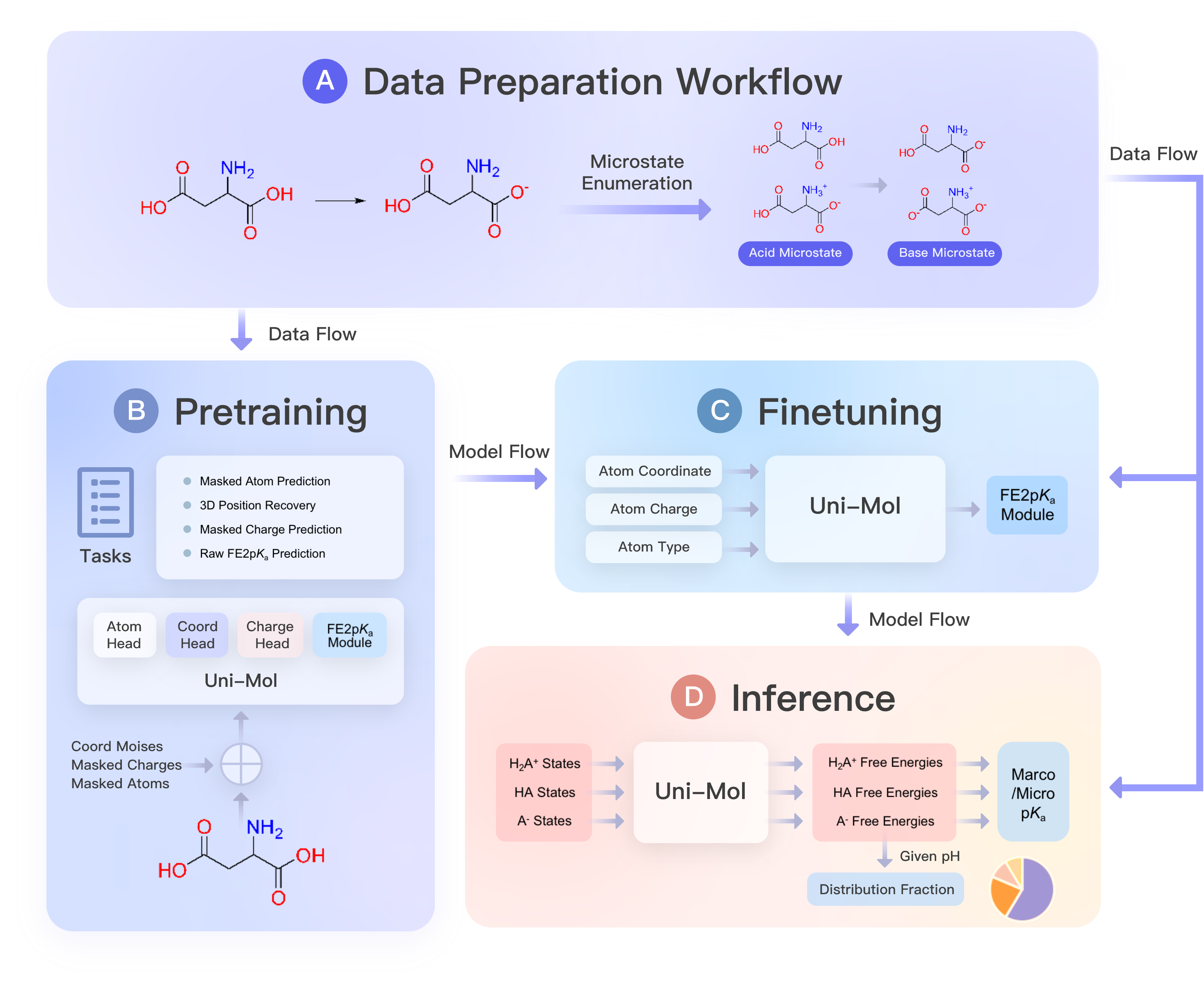
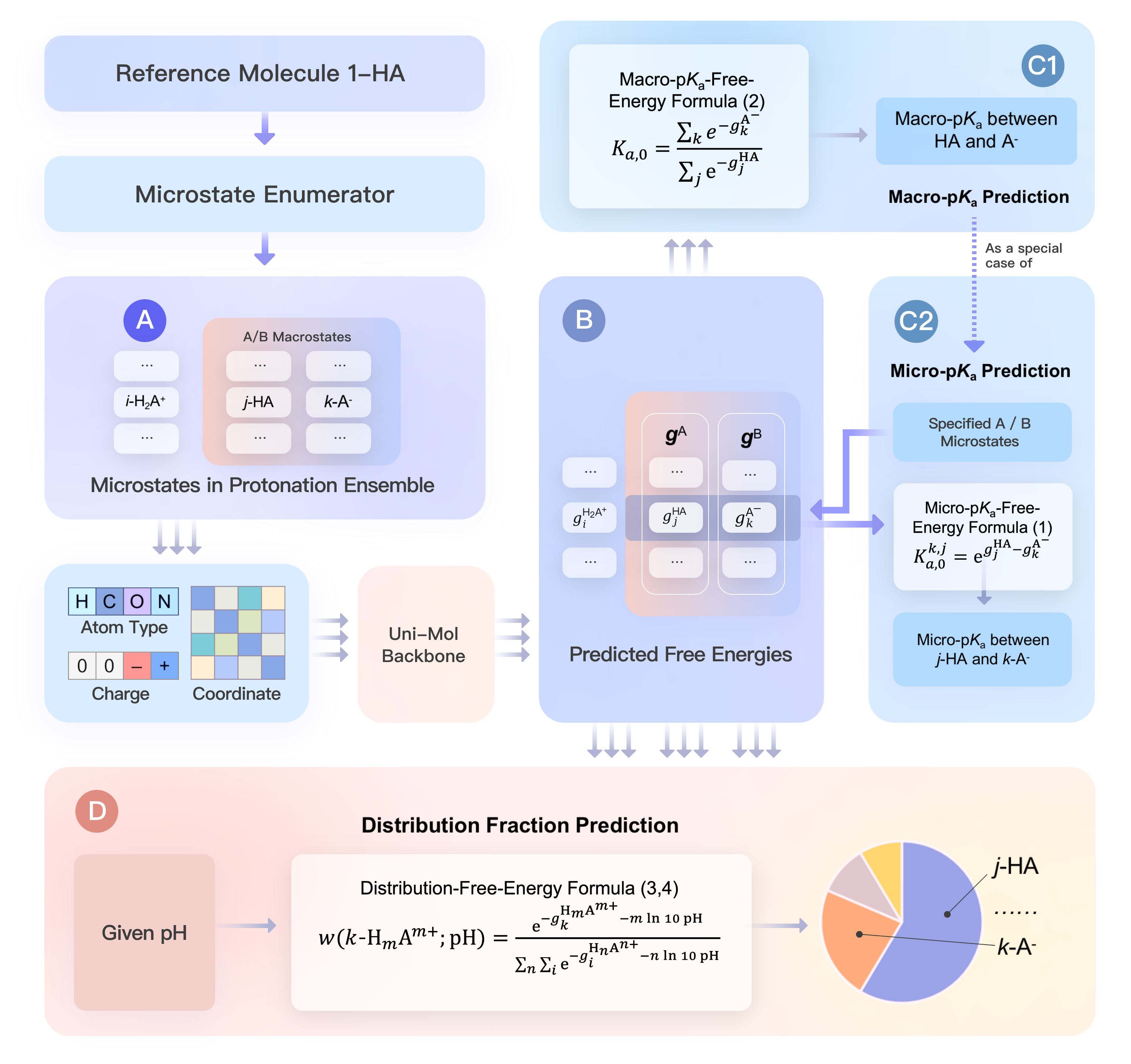
The official implementation of the model Uni-pKa in the paper Bridging Machine Learning and Thermodynamics for Accurate pKa Prediction.
Published paper at [JACS Au] Relevant preprint at [ChemRxiv] | Small molecule protonation state ranking demo at [Bohrium App] | Full datasets at [AISSquare]
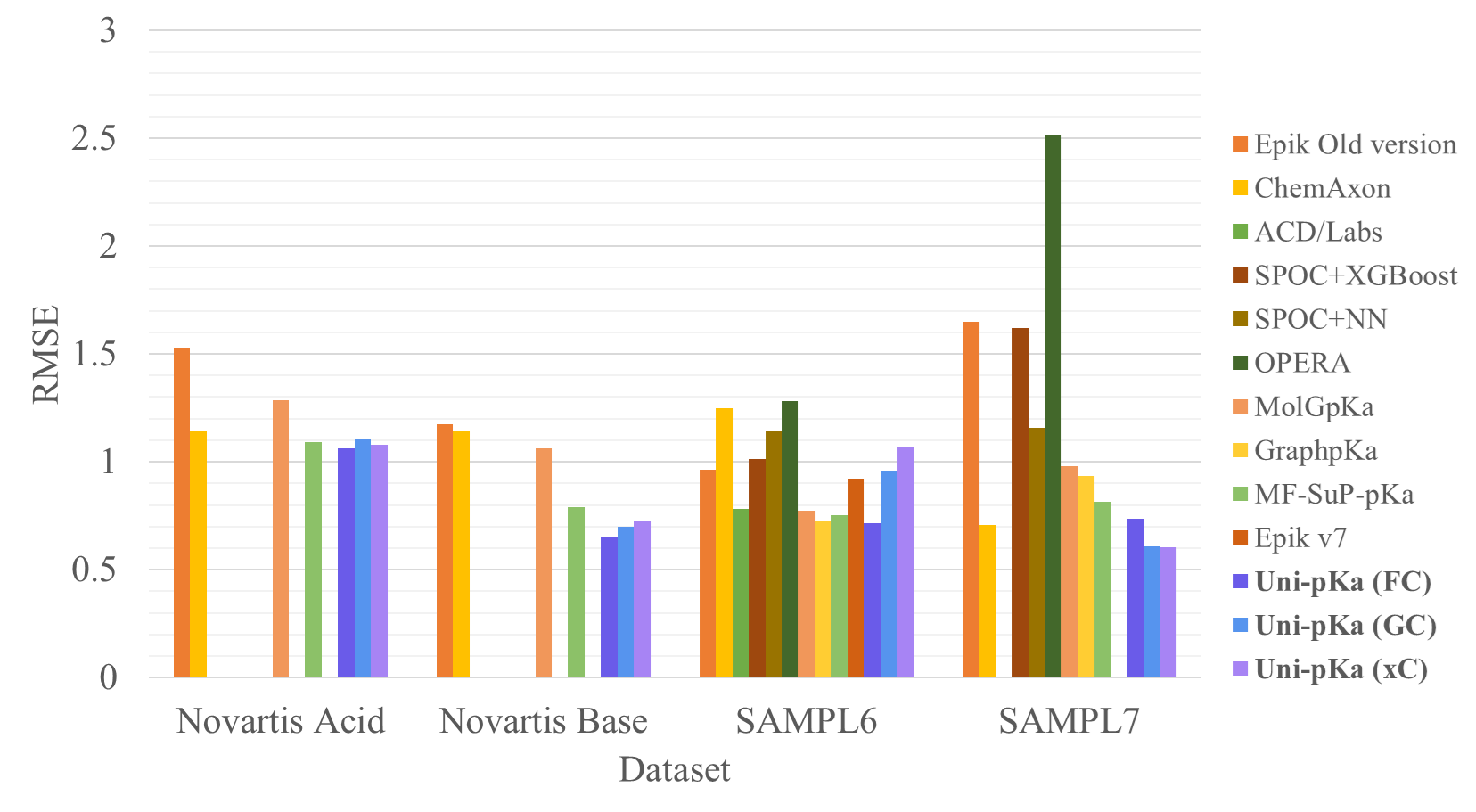
This machine-learning-based pKa prediction model achieves the state-of-the-art accuracy on several drug-like small molecule macro-pKa datasets.
Two core components of Uni-pKa framework are
-
A microstate enumerator to systematically build the protonation ensemble from a single structure.
-
A molecular machine learning model to predict the free energy of each single structure.
The model reaches the expected accuracy in the inference stage after the comprehensive data preparation by the enumerator, pretraining on the ChemBL dataset and finetuning on our Dwar-iBond dataset.
It uses iterated template-matching algorithm to enumerate all the microstates in adjacent macrostates of a molecule's protonation ensemble from at least one microstate stored as SMILES.
The protonation template smarts_pattern.tsv modifies and augments the one in the paper MolGpka: A Web Server for Small Molecule pKa Prediction Using a Graph-Convolutional Neural Network and its open source implementation (MIT license) in the Github repository MolGpKa.
The recommended environment is
python = 3.8.13
rdkit = 2021.09.5
numpy = 1.20.3
pandas = 1.5.2Reconstruct a plain pKa dataset to the Uni-pKa standard macro-pKa format with fully enumerated microstates
cd enumerator
python main.py reconstruct -i <input> -o <output> -m <mode>The <input> dataset is assumed be a csv-like file with a column storing SMILES. There are two cases allowed for each entry in the dataset.
- It contains only one SMILES. The Enumerator helps to build the protonated/deprotonated macrostate and complete the original macrostate.
- When
<mode>is "A", it will be considered as an acid (thrown into A pool). - When
<mode>is "B", it will be considered as a base (thrown into B pool).
- When
- It contains a string like "A1,...,Am>>B1,...Bn", where A1,...,Am are comma-separated SMILES of microstates in the acid macrostate (all thrown into A pool), and B1,...,Bn are comma-separated SMILES of microstates in the base macrostate(all thrown into B pool). The Enumerator helps to complete the both.
The <mode> "A" (default) or "B" determines which pool (A/B) is the reference structures and the starting point of the enumeration.
The <output> dataset is then constructed after the enumeration.
Example:
cd enumerator
python main.py ensemble -i ../dataset/sampl6.tsv -o example_out.tsv -u 2 -l -2 -t simple_smarts_pattern.tsvThe input dataset is SAMPL6 dataset as example. Reconstructed pKa dataset, or just any molecular dataset with an "SMILES" column with single molecular SMILES is supported as the input. In the output file, like example_out.tsv, columns include the original SMILES, and macrostates of total charge between the upper bound set by -u (default +2) and the lower bound set by -l (default -2). A simpler template is prepared as simple_smarts_pattern.tsv here for cleaner protonation ensembles which discard some rare structure motifs in the aqueous solution.
It is a Uni-Mol-based neural network. By embedding the neural network into thermodynamic relationship between the free energy and pKa throughout the training and inference stages, the framework preserves physical consistency and adapts to multiple tasks.
The dependencies of Uni-pKa are the same as those of Uni-Mol.
- Uni-Core, check its Installation Documentation.
- rdkit==2022.9.3, install via
pip install rdkit-pypi==2022.9.3
The recommended environment is the docker image.
docker pull dptechnology/unimol:latest-pytorch1.11.0-cuda11.3
See details in Uni-Mol repository.
The raw data can be downloaded from [AISSquare].
First, preprocess the ChemBL training and validation sets, and then pretrain the model:
# Preprocess training set
python ./scripts/preprocess_pka.py --raw-csv-file Datasets/tsv/chembl_train.tsv --processed-lmdb-dir chembl --task-name train
# Preprocess validation set
python ./scripts/preprocess_pka.py --raw-csv-file Datasets/tsv/chembl_valid.tsv --processed-lmdb-dir chembl --task-name valid
# Copy the necessary dict file
cp -r unimol/examples/* chembl
# Pretrain the model
bash pretrain_pka.shNote: The head_name in the subsequent scripts must match the task_name in pretrain_pka.sh.
Next, preprocess the dwar-iBond dataset and finetune the model:
# Preprocess
python ./scripts/preprocess_pka.py --raw-csv-file Datasets/tsv/dwar-iBond.tsv --processed-lmdb-dir dwar --task-name dwar-iBond
# Copy the necessary dict file
cp -r unimol/examples/* dwar
# Finetune the model
bash finetune_pka.shInfer with the finetuned model, taking novartis_acid as an example:
# Preprocess
python ./scripts/preprocess_pka.py --raw-csv-file Datasets/tsv/novartis_acid.tsv --processed-lmdb-dir novartis_acid --task-name novartis_acid
# Copy the necessary examples from unimol
cp -r unimol/examples/* novartis_acid
# Run inference
bash infer_pka.shTo test with other external test datasets, it may be necessary to modify data_path, infer_task, and results_path in infer_pka.sh.
After inference, extract the results to CSV files and calculate the performance metrics (e.g., MAE, RMSE) on the results:
python ./scripts/infer_mean_ensemble.py --task pka --nfolds 5 --results-path novartis_acid_resultsThe metrics are calculated using the average of the 5-fold model predictions.