GenomeSpy is a Python wrapper for the JavaScript visualization grammar library GenomeSpy. It enables the creation of interactive, GPU-accelerated genomic visualizations through a declarative grammar.
The Python package provides a high-level interface to create GenomeSpy visualizations. Under the hood, it:
- Generates JSON specifications that GenomeSpy can interpret
- Creates a temporary HTTP server to serve local data files
- Handles rendering in Jupyter notebooks by creating temporary HTML files
- Supports exporting standalone HTML files for sharing (note: referenced data files must remain accessible)
The javascript (official) version of GenomeSpy is developed in The Systems Biology of Drug Resistance in Cancer group at the University of Helsinki by Kari Lavikka. Original repo is at genome-spy/genome-spy.
Note: This package has been primarily tested with the hg38 (GRCh38) human genome assembly. While it may work with other assemblies, full compatibility is not guaranteed. PRs are welcomed!
- 🚀 GPU-accelerated rendering for fluid interaction with large datasets
- 🧬 Native support for genomic data formats including BigWig, BigBed, and Indexed FASTA
- 📊 Declarative visualization grammar for mapping data to visual channels
- 🔍 Interactive zooming and navigation for exploring genomic regions
- 📱 Responsive design that adapts to different screen sizes
- 🔧 Highly customizable with extensive configuration options
pip install genomespyHere's a simple example to get you started:
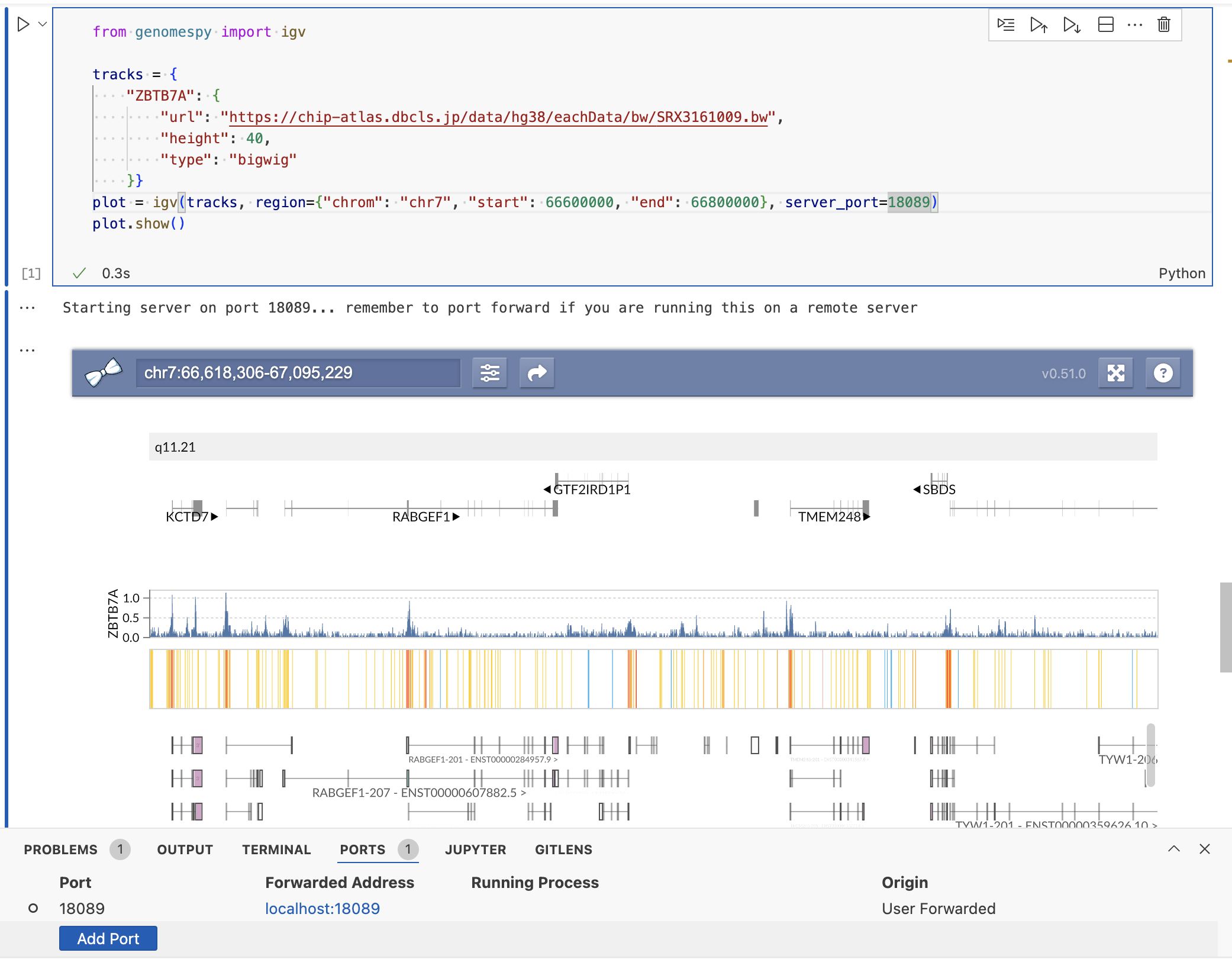
from genomespy import igv
tracks = {
"ZBTB7A": {
"url": "https://chip-atlas.dbcls.jp/data/hg38/eachData/bw/SRX3161009.bw",
"height": 40,
"type": "bigwig"
}}
plot = igv(tracks, region={"chrom": "chr7", "start": 66600000, "end": 66800000}, server_port=18089)
plot.show()
# plot.close() when you finished visualizationNote: When running on a remote server, you'll need to set up port forwarding to access the visualization locally. The default port is 18089, but this can be customized. In VS Code, you can set up port forwarding by:
- Opening the "PORTS" tab in the bottom panel
- Clicking "Forward a Port"
- Entering port 18089 (or your chosen port)
For other IDEs/environments, consult their documentation on port forwarding.
You can also use genomespy with gradio. Check out the example in example_gradio.py.
Comprehensive documentation is available at genomespy.readthedocs.io, including:
- Detailed installation instructions
- API reference
- Usage examples
- Contributing guidelines
GenomeSpy uses a grammar of graphics approach where visualizations are built by:
- Marks: Basic visual elements (rect, point, line, etc.)
- Encodings: Mappings from data to visual properties
- Scales: Functions that transform data values to visual values
- Transforms: Data transformations and calculations
- Parameters: Dynamic variables for interaction
To contribute to GenomeSpy:
# Clone the repository
git clone https://github.com/fuxialexander/genomespy.git
# Install development dependencies
cd genomespy
pip install -e ".[dev]"
# Run tests
pytestgenomespy is distributed under the terms of the MIT license.