Gene cluster comparison figure generator
clinker is a pipeline for easily generating publication-quality gene cluster comparison figures.
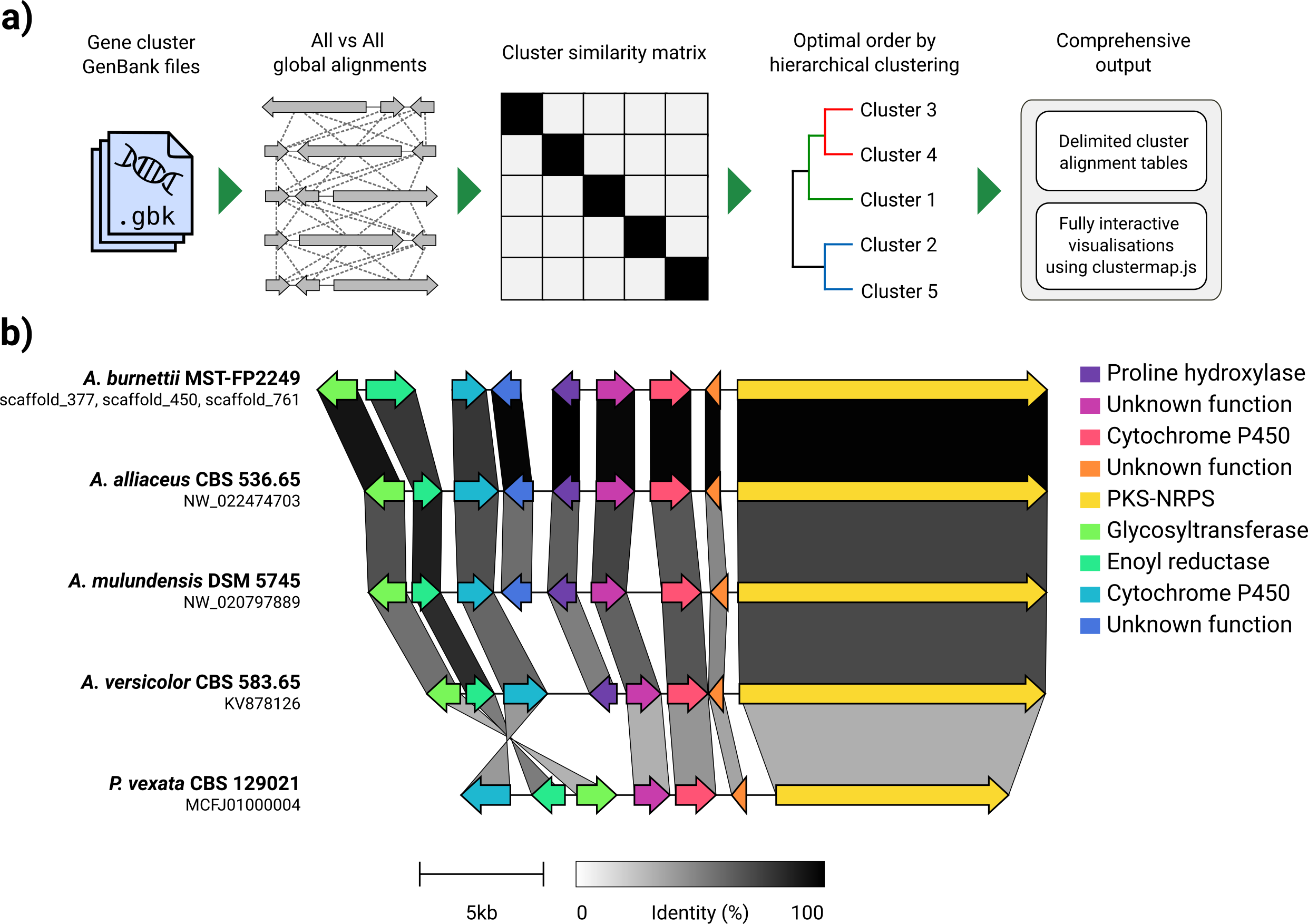
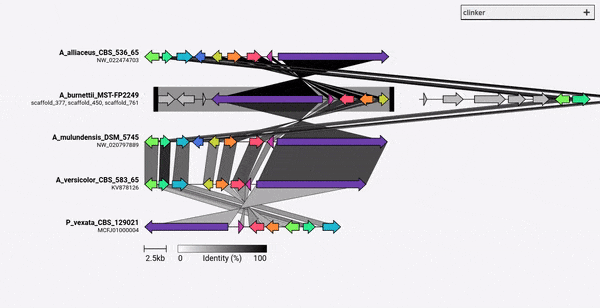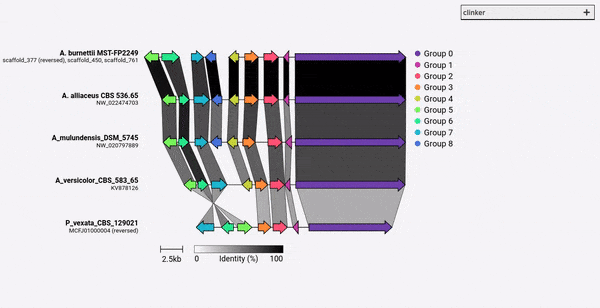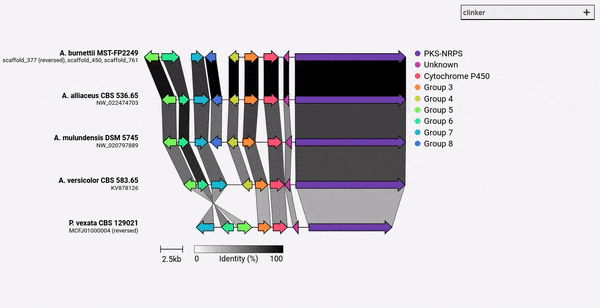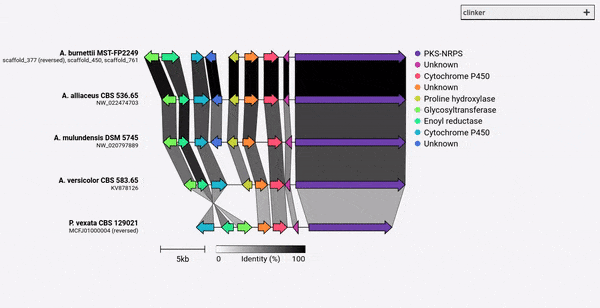
Given a set of GenBank files, clinker will automatically extract protein translations, perform global alignments between sequences in each cluster, determine the optimal display order based on cluster similarity, and generate an interactive visualisation (using clustermap.js) that can be extensively tweaked before being exported as an SVG file.
clinker can be installed directly through pip:
pip install clinker
By cloning the source code from GitHub:
git clone https://github.com/gamcil/clinker.git
cd clinker
pip install .
Or, through conda:
conda create -n clinker -c conda-forge -c bioconda clinker-py
conda activate clinker
If you found clinker useful, please cite:
clinker & clustermap.js: Automatic generation of gene cluster comparison figures.
Gilchrist, C.L.M., Chooi, Y.-H., 2020.
Bioinformatics. doi: https://doi.org/10.1093/bioinformatics/btab007
Running clinker can be as simple as:
clinker clusters/*.gbk
This will read in all GenBank files inside the folder, align them, and print
the alignments to the terminal. To generate the visualisation, use the -p/--plot
argument:
clinker clusters/*.gbk -p <optional: file name to save static HTML>
See -h/--help for more information:
usage: clinker [-h] [-i IDENTITY] [-f] [-o OUTPUT] [-dl DELIMITER]
[-dc DECIMALS] [-hl] [-ha]
files [files ...]
clinker: Automatic creation of publication-ready gene cluster comparison figures.
clinker generates gene cluster comparison figures from GenBank files.
It performs pairwise local or global alignments between every sequence
in every unique pair of clusters and generates interactive, to-scale
comparison figures using the clustermap.js library.
positional arguments:
files Gene cluster GenBank files
optional arguments:
-h, --help show this help message and exit
Alignment options:
-na, --no_align Do not align clusters
-i IDENTITY, --identity IDENTITY
Minimum alignment sequence identity
-j JOBS, --jobs JOBS Number of alignments to run in parallel (0 to use the
number of CPUs)
Output options:
-s SESSION, --session SESSION
Path to clinker session
-ji JSON_INDENT, --json_indent JSON_INDENT
Number of spaces to indent JSON
-f, --force Overwrite previous output file
-o OUTPUT, --output OUTPUT
Save alignments to file
-p [PLOT], --plot [PLOT]
Plot cluster alignments using clustermap.js. If a path
is given, clinker will generate a portable HTML file
at that path. Otherwise, the plot will be served
dynamically using Python's HTTP server.
-dl DELIMITER, --delimiter DELIMITER
Character to delimit output by
-dc DECIMALS, --decimals DECIMALS
Number of decimal places in output
-hl, --hide_link_headers
Hide alignment column headers
-ha, --hide_aln_headers
Hide alignment cluster name headers
Visualisation options:
-ufo, --use_file_order
Display clusters in order of input files
Example usage
-------------
Align clusters, plot results and print scores to screen:
$ clinker files/*.gbk -p
Cameron Gilchrist, 2020