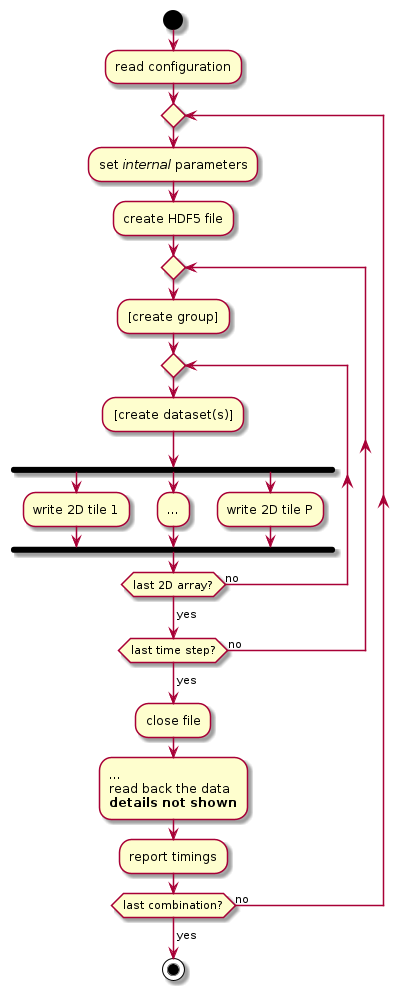
This is a simple I/O performance tester for HDF5. (See also ADIOS IOTEST.) Its purpose is to assess the performance variability of a set of logically equivalent HDF5 representations of a common pattern. The test repeatedly writes (and reads) in parallel a set of 2D array variables in a tiled fashion, over a set of “time steps.” A schematic is shown below.
A configuration file (see below) is used to control parameters such as the number of steps, the count and shape of the 2D array variables, etc. (See section sec:parameters.) The test then uses between 48 and 192 different combinations of up to 7 internal parameters (see section sec:internal-parameters) to configure the HDF5 library and output, to perform write and read operations, and to report certain timings. (The number of combinations is different for sequential and parallel runs, and depends on other choices.)
For a baseline, this test can (and should!) be run with a single MPI process using the POSIX, core, and MPI-IO VFDs. Typically, each baseline variant covers a different aspect of the underlying system, its configuration, and the particular HDF5 library version. Doing MPI-parallel runs without this baseline is the proverbial “getting off on the wrong foot.”
While it can be built manually, we recommend building hdf5_iotest from Spack.
- Create a new Spack package
spack create --name hdf5iotestThis creates a boilerplate
package.pyfile, which we’ll need to edit. - Edit the freshly minted
package.pyviaspack edit hdf5iotestpackage.pyshould look like this:# Copyright 2013-2020 Lawrence Livermore National Security, LLC and other # Spack Project Developers. See the top-level COPYRIGHT file for details. # # SPDX-License-Identifier: (Apache-2.0 OR MIT) from spack import * class Hdf5iotest(AutotoolsPackage): """ A simple I/O performance test for HDF5. Run with 'hdf5_iotest <INI file>'. A sample INI file called hdf5_iotest.ini can be found in the share/hdf5-iotest subdirectory. It is recommended to run multiple configurations. The combinator.sh script in share/hdf5-iotest creates 24 "standard" parameter cominations. """ homepage = "https://github.com/HDFGroup/hdf5-iotest" git = "https://github.com/HDFGroup/hdf5-iotest.git" maintainers = ['gheber'] version('master', branch='master') depends_on('autoconf', type='build') depends_on('automake', type='build') depends_on('libtool', type='build') depends_on('m4', type='build') depends_on('mpi') depends_on('hdf5') def configure_args(self): args = [] if "+gperftools" not in self.spec: args += ["--enable-gperftools"] return args def test(self): pass
- Test the configuration via
spack spec hdf5iotestSample output can be found in the appendix.
You need a working installation of parallel HDF5 (which depends on MPI).
./autogen.sh
CC=h5pcc ./configure --prefix=<installation directory>
make
make installhdf5_iotest accepts a single argument, the name of a configuration file. If no
configuration file name is provided, it looks for a configuration named
hdf5_iotest.ini in the current working directory.
hdf5_iotest [INI file]The configuration file syntax is shown below:
[DEFAULT]
<<hdf5-iotest-conf>>
A complete configuration can be found here.
Rather than modifying the [DEFAULT] section, we recommend that testers create
a new section, e.g., [CUSTOM], and overwrite the default values as needed.
[DEFAULT] ... [CUSTOM] ...
Any parameter specified in the [CUSTOM] section overwrites its [DEFAULT]
counterpart.
The following configuration parameters are supported.
- Version
- The HDF5 I/O test configuration version
version = 0Currently, 0 is the only valid version.
- Steps
- The number of steps or repetitions, a positive integer.
steps = 20 - Number of 2D Array Variables
- The number of 2D array variables to be
written, a positive integer.
arrays = 500 - Array Rows
- HDF5 I/O test can be run in strong or weak scaling mode (see
below). For strong scaling, this is the total number (across all MPI ranks)
of rows of each 2D array variable. For weak scaling, this is the number of
rows per MPI process per 2D array variable.
rows = 100 - Array Columns
- HDF5 I/O test can be run in strong or weak scaling mode
(see below). For strong scaling, this is the total number (across all MPI
ranks) of columns of each 2D array variable. For weak scaling, this is the
number of columns per MPI process per 2D array variable.
columns = 200 - Number of MPI Process Rows
- HDF5 I/O test is run over a logical 2D grid
of MPI processes. This is the number of MPI process rows.
process-rows = 1For strong scaling, the
rowsmust be divisible byprocess-rows. - Number of MPI Process Columns
- HDF5 I/O test is run over a logical 2D grid
of MPI processes. This is the number of MPI process columns.
process-columns = 1For strong scaling, the
columnsparameter must be divisible byprocess-columns. - Scaling<sec:scaling>
- HDF5 I/O test can be run with strong or weak
scaling. In strong scaling mode, the total amount of data written and read
is independent of the number of MPI processes, i.e., the per process I/O share
diminishes with an increase in the number of I/O processes. In weak scaling
mode, the amount of data written and read by each MPI-process is kept
constant, and the total I/O increases with the number of MPI processes.
# [weak, strong] scaling = weak - Alignment Increment
- Align HDF5 objects greater than or equal to an
alignment threshold on addresses which are a multiple of this increment.
# align along increment [bytes] boundaries alignment-increment = 1By default, there are no alignment restrictions in effect, and only increments greater than 1 have any effect.
- Alignment Threshold
- The minimum object size (in bytes) for which alignment
constraints will be enforced. A threshold of 0 forces everything to be
aligned.
# minimum object size [bytes] to force alignment (0=all objects) alignment-threshold = 0 - Meta Block Size
- The minimum size of metadata block allocations
when
H5FD_FEAT_AGGREGATE_METADATAis set by a VFL driver.# minimum metadata block allocation size in [bytes] meta-block-size = 2048Setting the value to 0 with this function will turn off metadata aggregation, even if the VFL driver attempts to use the metadata aggregation strategy.
The POSIX, core, and MPI-IO VFDs all support metadata allocation aggregation.
- Single Process I/O
- The I/O driver or mode to be used when running with a
single process.
# [posix, core, mpi-io-uni] single-process = posixThis setting is important when establishing a single-process baseline.
posixuses the default POSIX VFD.coreuses a memory-backed HDF5 file where the underlying memory buffer grows in 64 MB increments.mpi-io-uniuses the MPI-IO VFD (with a single process). - HDF5 Output File Name
- The default HDF5 output file name is
hdf5_iotest.h5. Use this parameter to select a different name. Note: The character “#” in the filename is reserved for creating an HDF5 with the “#” replaced with the accumulated case number, for example hdf5_iotest.#.h5.hdf5-file = hdf5_iotest.h5 - Results File
- When running the HDF5 I/O test, certain metrics are printed to
stdout. To simplify the analysis of results from multiple runs, they are also written to a CSV file whose name is configurable.csv-file = hdf5_iotest.csv - Restart
- The simulations will resume from (and including) the last successful
entry in the result’s CSV file. A value of 1 indicates a restart run,
and 0 is no restart. If the keyword is not present, the default is
not a restart.
# [0, 1] restart = 1 - Split
- The split I/O driver will be used [1]. Zero indicates to not use
the split file diriver, default.
# [0, 1] split = 1 - One-case
- The simulation will run only one case in the parameter space.
A value not equal to 0 indicates which parameter case to run, and the
value is a cumulative counter of the nested loops over the parameter space.
For example, to rerun the 100th case in the CSV file, the value would be 100.
If the keyword is not present, the default is to do all the cases.
# case number in the parameter space to run. one-case = 100 - Compression
- Specifies the compression filter for chunked datasets and
currently supports gzip and szip. The value corresponds to
parameters in the corresponding HDF5 API. Valid parameters for ”gzip” is an
integer for the level (see H5Pset_deflate). For ”szip” value is
options_mask, and pixels_per_block (see H5Pset_szip).
# compression filter (gzip, szip). szip = H5_SZIP_NN_OPTION_MASK, 8
Currently, the I/O test varies the following parameters:
- Dataset Rank
- The 2D array variables can be stored individually, or embedded into 3D or 4D datasets. In other words, the rank can be 2, 3, or 4.
- Slowest Dimension
- The slowest dimension can be array (count) or time.
- Initialization with Fill Values
- The default behavior of the HDF5 library is to initialize storage with the default or a user-specified fill value. This incurs additional I/O and may reduce performance.
- Storage Layout
- The dataset storage layout in the HDF5 file can be chunked or contiguous (or compact or virtual or user-defined).
- Alignment
- HDF5 objects greater than or equal to an alignment threshold can be aligned on addresses that are a multiple of a certain increment.
- Lower Library Version Bound
- The HDF5 library can be configured to use the earliest or latest available file format micro-versions when generating objects.
- MPI I/O Operations
- With MPI, the write and read operations can be collective or independent.
Since there is no shortage of knobs in the HDF5 API, other parameters might be added in the future.
==> Using specified package name: 'hdf5iotest' ==> Created template for hdf5iotest package ==> Created package file: /home/gerdheber/GitHub/spack/var/spack/repos/builtin/packages/hdf5iotest/package.py Waiting for Emacs... % spack spec hdf5iotest Input spec -------------------------------- hdf5iotest Concretized -------------------------------- hdf5iotest@spack%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected]+sigsegv patches=3877ab548f88597ab2327a2230ee048d2d07ace1062efe81fc92e91b7f39cd00,fc9b61654a3ba1a8d6cd78ce087e7c96366c290bc8d2c299f09828d793b853c8 arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected]+cpanm+shared+threads arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected]~symlinks+termlib arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected]~cxx~debug~fortran~hl~java+mpi+pic+shared~szip~threadsafe api=none arch=linux-debian10-skylake ^[email protected]%[email protected]~atomics~cuda~cxx~cxx_exceptions+gpfs~java~legacylaunchers~lustre~memchecker~pmi~singularity~sqlite3+static~thread_multiple+vt+wrapper-rpath fabrics=none schedulers=none arch=linux-debian10-skylake ^[email protected]%[email protected]~cairo~cuda~gl~libudev+libxml2~netloc~nvml+pci+shared arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected]~python arch=linux-debian10-skylake ^[email protected]%[email protected] arch=linux-debian10-skylake ^[email protected]%[email protected]~pic arch=linux-debian10-skylake ^[email protected]%[email protected]+optimize+pic+shared arch=linux-debian10-skylake ^[email protected]%[email protected] patches=4e1d78cbbb85de625bad28705e748856033eaafab92a66dffd383a3d7e00cc94 arch=linux-debian10-skylake
The CSV output looks like this. It’s then easy to concatenate several of these (after stripping out the header), and load them into pandas or R.
steps,arrays,rows,cols,scaling,proc-rows,proc-cols,slowdim,rank,version,alignment-increment,alignment-threshold,layout,fill,fmt,io,wall [s],fsize [B],write-phase-min [s],write-phase-max [s],creat-min [s],creat-max [s],write-min [s],write-max [s],read-phase-min [s],read-phase-max [s],read-min [s],read-max [s] 20,500,100,200,weak,1,1,step,2,"1.8.22",1,0,contiguous,true,earliest,core,15.37,1689710848,14.10,14.10,7.25,7.25,0.77,0.77,1.27,1.27,0.27,0.27 20,500,100,200,weak,1,1,step,2,"1.8.22",1,0,contiguous,true,latest,core,20.70,1682116757,19.53,19.53,12.08,12.08,0.78,0.78,1.17,1.17,0.28,0.28 ...
All timings are obtained via MPI_Wtime. For some metrics, we record minima and
maxima across MPI ranks. The columns steps through mpi-io are just
reiterations of the configuration parameters. The remaining columns are as
follows:
wall [s]- Wall time in seconds.
fsize [B]- The HDF5 output file size in bytes
write-phase-min [s],write-phase-max [s]- The fastest and slowest cumulative write phase time in seconds. This includes the time for file and dataset creation(s).
creat-min [s],creat-max [s]- The fastest and slowest time spent in
H5Fcreate,H5Fclose, andH5Dcreatein seconds write-min [s],write-max [s]- The fastest and slowest cumulative
H5Dwritetime in seconds read-phase-min [s],read-phase-max [s]- The fastest and slowest cumulative read phase time in seconds. This includes the times for opening and closing the HDF5 file and for creating dataset selections.
read-min [s],read-max [s]- The fastest and slowest cumulative
H5Dreadtime in seconds