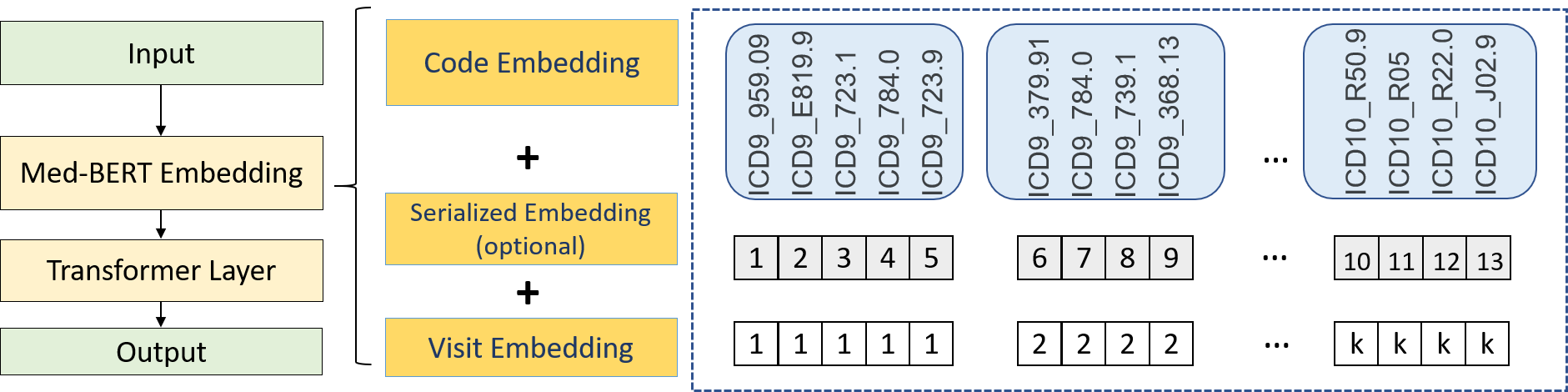
This repository provides the code for pre-training and fine-tuning Med-BERT, a contextualized embedding model that delivers a meaningful performance boost for real-world disease-prediction problems as compared to state-of-the-art models.
Med-Bert adapts bidirectional encoder representations from transformers (BERT) framework and pre-trains contextualized embeddings for diagnosis codes mainly in ICD-9 and ICD-10 format using structured data from an EHR dataset containing 28,490,650 patients.
 Please refer to our paper Med-BERT: pre-trained contextualized embeddings on large-scale structured electronic health records for disease prediction for more details.
Please refer to our paper Med-BERT: pre-trained contextualized embeddings on large-scale structured electronic health records for disease prediction for more details.
To reproduce the steps necessary for pre-training Med-BERT
python preprocess_pretrain_data.py <data_File> <vocab/NA> <output_Prefix> <subset_size/0forAll>
python create_BERTpretrain_EHRfeatures.py --input_file=<output_Prefix.bencs.train> --output_file='output_file' --vocab_file=<output_Prefix.types>--max_predictions_per_seq=1 --max_seq_length=64
python run_EHRpretraining.py --input_file='output_file' --output_dir=<path_to_outputfolder> --do_train=True --do_eval=True --bert_config_file=config.json --train_batch_size=32 --max_seq_length=512 --max_predictions_per_seq=1 --num_train_steps=4500000 --num_warmup_steps=10000 --learning_rate=5e-5
You can find an example for the construction of the data_file under Example data as well as images showing the construction of preprocessed data and the BERT features. Additional details are available under Pretraining Tutorial
Note: We run our code using mainly GPU, while CPU and TPU options migt be available in the code they were not tested.
To see an example of how to use Med-BERT for a specific disease prediction task, you can follow the Med-BERT DHF prediction notebook
Kindly note that you need to use the following code for preparing the fine-tunning data using (create_ehr_pretrain_FTdata.py) in a similar way of preparing the pretraining data.
Python: 3.7+
Pytorch 1.5.0
Tensorflow 1.13.1+
Pandas
Pickle
tqdm
pytorch-transformers
Google BERT
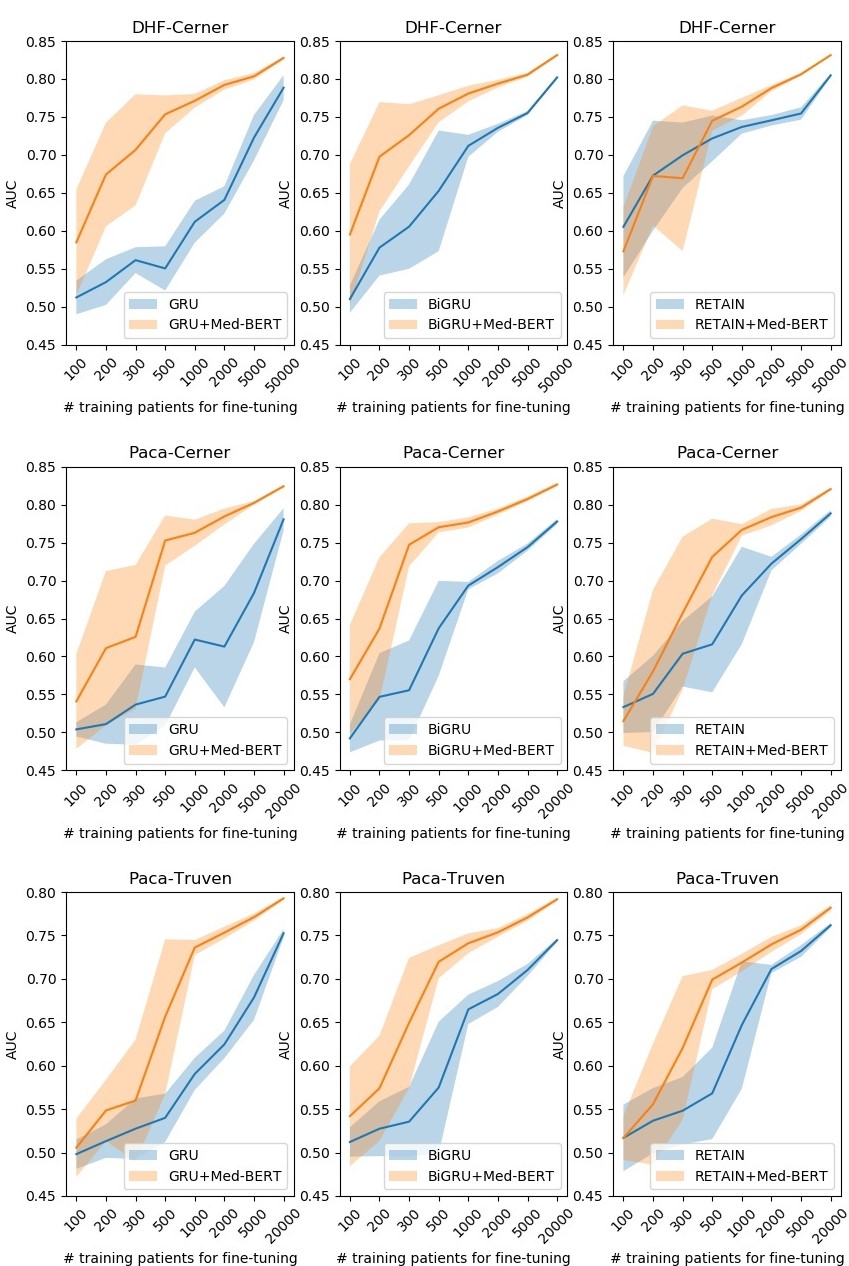
Prediction results for the evaluation sets by training on different sizes of data on DHF-Cerner (top), PaCa-Cerner (middle), and PaCa-Truven (bottom). The shadows indicate the standard deviations. Please refer to our paper for more details.
Initially we really hoped to share our models but unfortunately, the pre-trained models are no longer sharable. According to SBMI Data Service Office: "Under the terms of our contracts with data vendors, we are not permitted to share any of the data utilized in our publications, as well as large models derived from those data."
Please post a Github issue if you have any questions.
Please acknowledge the following work in papers or derivative software:
Laila Rasmy, Yang Xiang, Ziqian Xie, Cui Tao, and Degui Zhi. "Med-BERT: pre-trained contextualized embeddings on large-scale structured electronic health records for disease prediction." npj digital medicine 2021 https://www.nature.com/articles/s41746-021-00455-y.