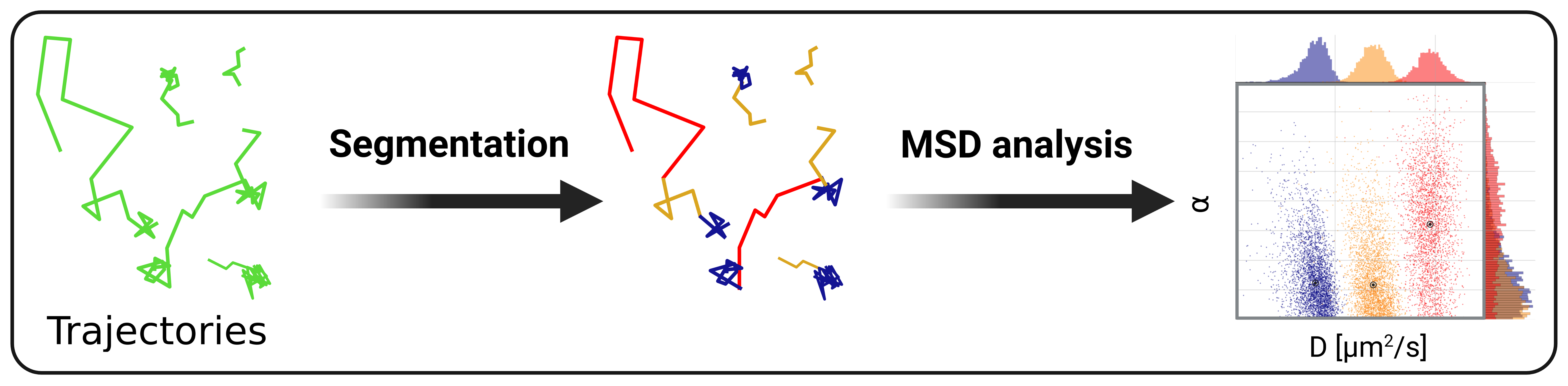
Trajectory segmentation into diffusive states using LSTM neural network.
git clone https://github.com/hkabbech/TrackSegNet.git
cd TrackSegNet
On Unix or MacOS, run:
source tracksegnet-env/bin/activate
On Windows, run:
tracksegnet-env\bin\activate
Note, to deactivate the virtual environment, type deactivate
pip3 install virtualenv
virtualenv -p /usr/bin/python3 tracksegnet-env
source tracksegnet-env/bin/activate # for Windows: tracksegnet-env\bin\activate
python -m pip install -r requirements.txt
Organize your data in a folder SPT_experiment, each sub-folder should contain a file storing the trajectory coordinates in a MDF or CSV file format.
If CSV format is used, the headers should be: x, y, frame, track_id
.
├── data/
│ └── SPT_experiment/
│ ├── Cell_1
│ │ ├── *.tif
│ │ └── *.mdf
│ ├── Cell_2
│ │ ├── *.tif
│ │ └── *.mdf
│ ├── Cell_3
│ │ ├── *.tif
│ │ └── *.mdf
│ └── ...
│
├── src/
├── tracksegnet-env/
├── parms.csv
├── tracksegnet.py
└── ...
Update the main parameters in the parms.csv file according to your experiment:
-
data_path: the path containing your data folderSPT_experimentto analyze -
track_format: The format of the files containing the trajectory coordinates, should beMDForCSV -
time_frame: the time interval between two trajectory points (in second) -
pixel_size: the dimension of a pixel (in µm) -
num_states: the number of diffusive states for the classification(from 2 to 6 states) -
state_X_diff: The expected diffusion value for state X (in µm^2/s). -
state_X_alpha: The expected anomalous exponent α value for state X (from 0 to 2 -- ]0-1[: subdiffusion, 1: Brownian motion, ]1-2[: superdiffusion). -
pt_i_j: the probability of transitionning from the state i to the state j. The total number of probabilities should be$N^2$ .
Note that the program will run on the toy example if the parameters are unchanged.
For updating the parameters of the track simulation and neural network training, please make the changes in the main file tracksegnet.py.
./tracksegnet.py parms.csv
Arts, M., Smal, I., Paul, M. W., Wyman, C., & Meijering, E. (2019). Particle Mobility Analysis Using Deep Learning and the Moment Scaling Spectrum. Scientific Reports, 9(1), 17160. https://doi.org/10.1038/s41598-019-53663-8.