PROJECT NOT UNDER ACTIVE MANAGEMENT
This project will no longer be maintained by Intel.
Intel has ceased development and contributions including, but not limited to, maintenance, bug fixes, new releases, or updates, to this project.
Intel no longer accepts patches to this project.
If you have an ongoing need to use this project, are interested in independently developing it, or would like to maintain patches for the open source software community, please create your own fork of this project.
Contact: [email protected]
Many reference kits in the bio-medical domain focus on a single-model and single-modal solution. Exclusive reliance on a single method has some limitations, such as impairing the design of robust and accurate classifiers for complex datasets. To overcome these limitations, we provide this multi-modal disease prediction reference kit.
Multi-modal disease prediction is an Intel optimized, end-to-end reference kit for fine-tuning and inference. This reference kit implements a multi-model and multi-modal solution that will help to predict diagnosis by using categorized contrast enhanced mammography data and radiologists’ notes.
You can find a blog article related to this topic by following this link.
Check out more workflow and reference kit examples in the Developer Catalog.
- Solution Technical Overview
- Validated Hardware Details
- Software Requirements
- How it Works?
- Get Started
- Run Using Docker
- Run Using Argo Workflows on K8s using Helm
- Run Using Bare Metal
- Run Using Jupyter Lab
- Expected Output
- Summary and Next Steps
- Learn More
- Troubleshooting
- Support
This reference kit demonstrates one possible reference implementation of a multi-model and multi-modal solution. While the vision workflow aims to train an image classifier that takes in contrast-enhanced spectral mammography (CESM) images, the natural language processing (NLP) workflow aims to train a document classifier that takes in annotation notes about a patient’s symptoms. Each pipeline creates prediction for the diagnosis of breast cancer. In the end, weighted ensemble method is used to create final prediction.
The goal is to minimize an expert’s involvement in categorizing samples as normal, benign, or malignant, by developing and optimizing a decision support system that automatically categorizes the CESM with the help of radiologist notes.
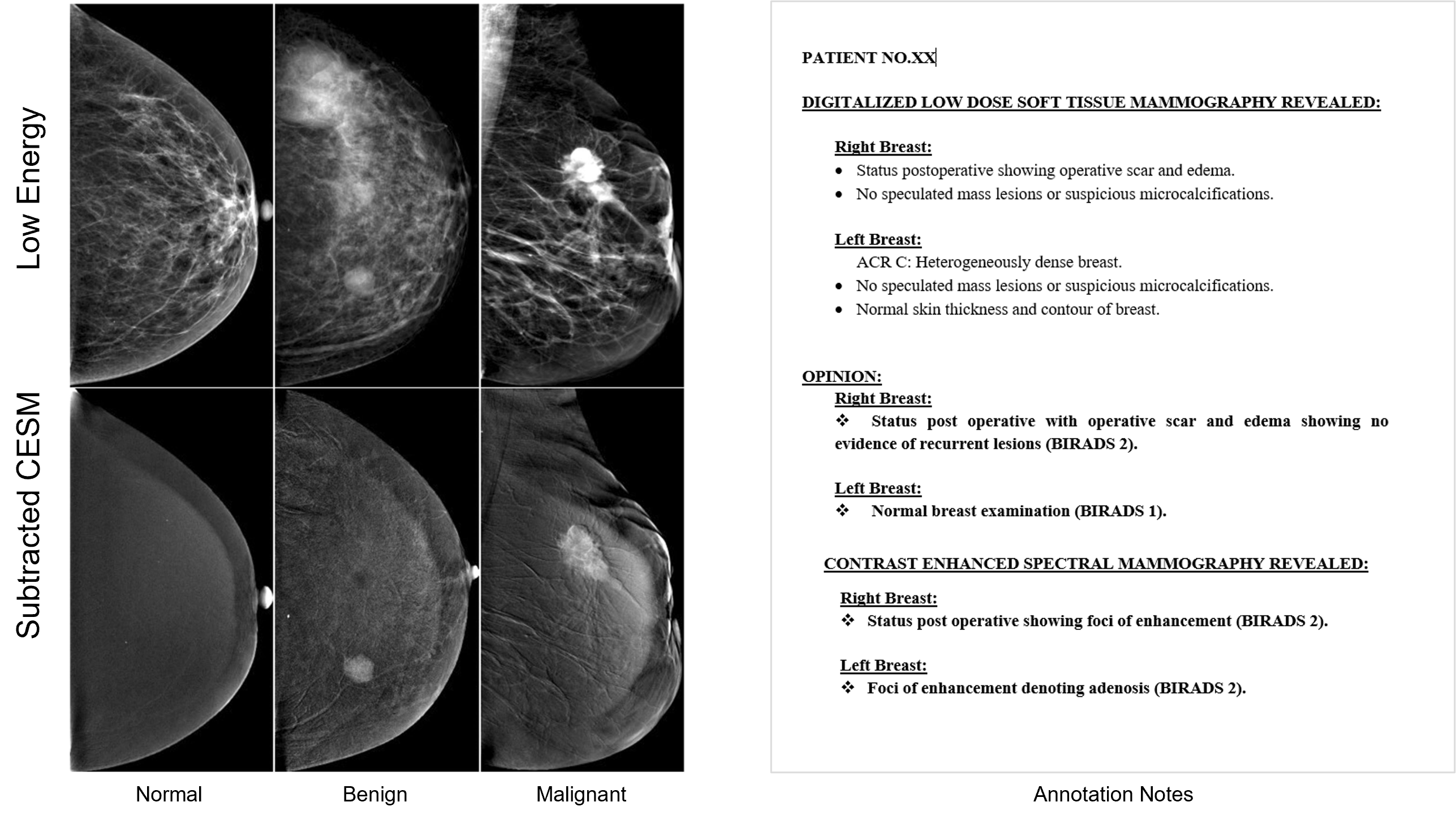
The dataset is a collection of 2,006 high-resolution contrast-enhanced spectral mammography (CESM) images (1003 low energy images and 1003 subtracted CESM images) with annotations of 326 female patients. See Figure-1. Each patient has 8 images, 4 representing each side with two views (Top Down looking and Angled Top View) consisting of low energy and subtracted CESM images. Medical reports, written by radiologists, are provided for each case along with manual segmentation annotation for the abnormal findings in each image. As a preprocessing step, we segment the images based on the manual segmentation to get the region of interest and group annotation notes based on the subject and breast side.
Figure-2: Samples of low energy and subtracted CESM images and Medical reports, written by radiologists from the Categorized contrast enhanced mammography dataset. (Khaled, 2022)
For more details of the dataset, visit the wikipage of the CESM and read Categorized contrast enhanced mammography dataset for diagnostic and artificial intelligence research.
There are workflow-specific hardware and software setup requirements depending on how the workflow is run. Bare metal development system and Docker image running locally have the same system requirements.
| Recommended Hardware | Precision |
|---|---|
| Intel® 4th Gen Xeon® Scalable Performance processors | FP32, BF16 |
| Intel® 1st, 2nd, 3rd, and 4th Gen Xeon® Scalable Performance processors | FP32 |
To execute the reference solution presented here, use CPU for fine tuning.
Linux OS (Ubuntu 22.04) is used to validate this reference solution. Make sure the following dependencies are installed.
sudo apt-get updatesudo apt-get install -y build-essential gcc git libgl1-mesa-glx libglib2.0-0 python3-devsudo apt-get install -y python3.9 python3-pip, and some virtualenv like python3-venv or condapip install dataset-librarian
Note: The current version of dataset-librarian requires Python version 3.9 or later, up to version 3.11. If your current Python version does not meet this requirement, please proceed with Set Up System Software first.
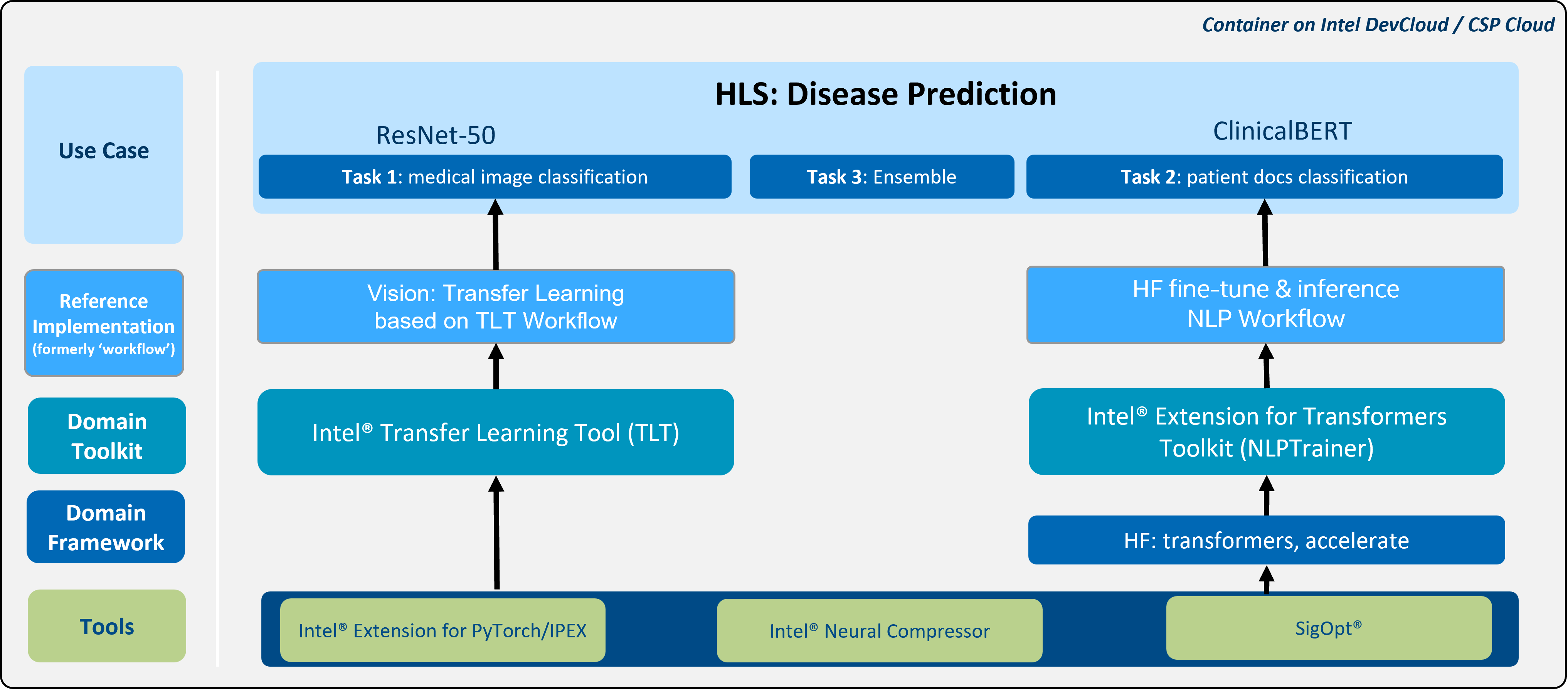 Figure-1: Architecture of the reference kit
Figure-1: Architecture of the reference kit
- Uses real-world CESM breast cancer datasets with “multi-modal and multi-model” approaches.
- Two domain toolkits (Intel® Transfer Learning Toolkit and Intel® Extension for Transformers), Intel® Neural Compressor and other libs/tools and uses Hugging Face model repo and APIs for ResNet-50 and ClinicalBert models.
- The NLP reference Implementation component uses HF Fine-tuning and Inference Optimization workload, which is optimized for document classification. This NLP workload employs Intel® Neural Compressor and other libraries/tools and utilizes Hugging Face model repository and APIs for ClinicalBert models. The ClinicalBert model, which is pretrained with a Masked-Language-Modeling task on a large corpus of English language from MIMIC-III data, is fine-tuned with the CESM breast cancer annotation dataset to generate a new BERT model.
- The Vision reference Implementation component uses TLT-based vision workload, which is optimized for image fine-tuning and inference. This workload utilizes Intel® Transfer Learning Tool and tfhub's ResNet-50 model to fine-tune a new convolutional neural network model with subtracted CESM image dataset. The images are preprocessed by using domain expert-defined segmented regions to reduce redundancies during training.
- Predict diagnosis by using categorized contrast enhanced mammography images and radiologists’ notes separately and weighted ensemble method applied to results of sub-models to create the final prediction.
Start by defining an environment variable that will store the workspace path, this can be an existing directory or one to be created in further steps. This ENVVAR will be used for all the commands executed using absolute paths.
E. g.
export WORKSPACE=/mydisk/mtw/myworkCreate a working directory for the reference kit and clone the Breast Cancer Prediction Reference Kit repository into your working directory.
git clone https://github.com/intel/disease-prediction.git $WORKSPACE/brca_multimodal
cd $WORKSPACE/brca_multimodal
git submodule update --init --recursive
Use the links below to download the image datasets. Or skip to the Docker section to download the dataset using a container.
Once you've downloaded the image file (which will be in zip format) and placed it in the data directory, unzip it using the following command.
cd data
unzip CDD-CESM.zip
After that, proceed by executing the following command. This will initiate the download of segmentation and annotation data, followed by segmentation and preprocessing operations.
Command-line Interface:
- -d : Directory location where the raw dataset will be saved on your system. It's also where the preprocessed dataset files will be written. If not set, a directory with the dataset name will be created.
- --split_ratio: Split ratio of the test data, the default value is 0.1.
More details of the dataset_librarian can be found here.
cd $WORKSPACE/brca_multimodal
python -m dataset_librarian.dataset -n brca --download --preprocess -d data/ --split_ratio 0.1
Note: See this dataset's applicable license for terms and conditions. Intel Corporation does not own the rights to this dataset and does not confer any rights to it.
This reference kit offers three options for running the fine-tuning and inference processes:
- Docker
- Argo Workflows on K8s Using Helm
- Bare Metal
- Jupyter Workspace
Details about each of these methods can be found below.
Follow these instructions to set up and run our provided Docker image. For running on bare metal, see the bare metal instructions.
You'll need to install Docker Engine on your development system. Note that while Docker Engine is free to use, Docker Desktop may require you to purchase a license. See the Docker Engine Server installation instructions for details.
If the Docker image is run on a cloud service, mention they may also need credentials to perform training and inference related operations (such as these for Azure):
- Set up the Azure Machine Learning Account
- Configure the Azure credentials using the Command-Line Interface
- Compute targets in Azure Machine Learning
- Virtual Machine Products Available in Your Region
To build and run this workload inside a Docker Container, ensure you have Docker Compose installed on your machine. If you don't have this tool installed, consult the official Docker Compose installation documentation.
DOCKER_CONFIG=${DOCKER_CONFIG:-$HOME/.docker}
mkdir -p $DOCKER_CONFIG/cli-plugins
curl -SL https://github.com/docker/compose/releases/download/v2.7.0/docker-compose-linux-x86_64 -o $DOCKER_CONFIG/cli-plugins/docker-compose
chmod +x $DOCKER_CONFIG/cli-plugins/docker-compose
docker compose versionEnter the docker directory.
cd $WORKSPACE/brca_multimodal/dockerBuild or Pull the provided docker image.
docker compose build hf-nlp-wf vision-tlt-wf
docker compose build devOR
docker pull intel/ai-workflows:pa-hf-nlp-disease-prediction
docker pull intel/ai-workflows:pa-vision-tlt-disease-prediction
docker pull intel/ai-workflows:pa-disease-predictionPrepare dataset for Disease Prediction workflows and accept the legal agreement to use the Intel Dataset Downloader.
USER_CONSENT=y docker compose run preprocess| Environment Variable Name | Default Value | Description |
|---|---|---|
| DATASET_DIR | $PWD/../data |
Unpreprocessed dataset directory |
| SPLIT_RATIO | 0.1 |
Train/Test Split Ratio |
| USER_CONSENT | n/a | Consent to legal agreement |
Both NLP and Vision Fine-tuning containers must complete successfully before the Inference container can begin. The Inference container uses checkpoint files created by both the nlp and vision fine-tuning containers stored in the ${OUTPUT_DIR} directory to complete inferencing tasks.
%%{init: {'theme': 'dark'}}%%
flowchart RL
VDATASETDIR{{"/${DATASET_DIR"}} x-. "-$PWD/../data}" .-x hf-nlp-wf[hf-nlp-wf]
VCONFIGDIR{{"/${CONFIG_DIR"}} x-. "-$PWD/../configs}" .-x hf-nlp-wf
VOUTPUTDIR{{"/${OUTPUT_DIR"}} x-. "-$PWD/../output}" .-x hf-nlp-wf
VDATASETDIR x-. "-$PWD/../data}" .-x vision-tlt-wf[vision-tlt-wf]
VCONFIGDIR x-. "-$PWD/../configs}" .-x vision-tlt-wf
VOUTPUTDIR x-. "-$PWD/../output}" .-x vision-tlt-wf
VDATASETDIR x-. "-$PWD/../data}" .-x ensemble-inference[ensemble-inference]
VCONFIGDIR x-. "-$PWD/../configs}" .-x ensemble-inference
VOUTPUTDIR x-. "-$PWD/../output}" .-x ensemble-inference
ensemble-inference --> hf-nlp-wf
ensemble-inference --> vision-tlt-wf
classDef volumes fill:#0f544e,stroke:#23968b
class VDATASETDIR,VCONFIGDIR,VOUTPUTDIR,VDATASETDIR,VCONFIGDIR,VOUTPUTDIR,VDATASETDIR,VCONFIGDIR,VOUTPUTDIR volumes
Run entire pipeline to view the logs of different running containers.
docker compose run ensemble-inference &| Environment Variable Name | Default Value | Description |
|---|---|---|
| CONFIG | disease_prediction_container |
Config file name |
| CONFIG_DIR | $PWD/../configs |
Disease Prediction Configurations directory |
| DATASET_DIR | $PWD/../data |
Preprocessed dataset directory |
| OUTPUT_DIR | $PWD/../output |
Logfile and Checkpoint output |
Follow logs of each individual pipeline step using the commands below:
docker compose logs vision-tlt-wf -f
docker compose logs hf-nlp-wf -fTo view inference logs
fgCreate your own script and run your changes inside of the container or run inference without waiting for fine-tuning.
%%{init: {'theme': 'dark'}}%%
flowchart RL
VDATASETDIR{{"/${DATASET_DIR"}} x-. "-$PWD/../data}" .-x dev
VCONFIGDIR{{"/${CONFIG_DIR"}} x-. "-$PWD/../configs}" .-x dev
VOUTPUTDIR{{"/${OUTPUT_DIR"}} x-. "-$PWD/../output}" .-x dev
classDef volumes fill:#0f544e,stroke:#23968b
class VDATASETDIR,VCONFIGDIR,VOUTPUTDIR volumes
Run using Docker Compose.
docker compose run dev| Environment Variable Name | Default Value | Description |
|---|---|---|
| CONFIG | disease_prediction_container |
Config file name |
| CONFIG_DIR | $PWD/../configs |
Disease Prediction Configurations directory |
| DATASET_DIR | $PWD/../data |
Preprocessed Dataset |
| OUTPUT_DIR | $PWD/output |
Logfile and Checkpoint output |
| SCRIPT | src/breast_cancer_prediction.py |
Name of Script |
If your environment requires a proxy to access the internet, export your development system's proxy settings to the docker environment:
export DOCKER_RUN_ENVS="-e ftp_proxy=${ftp_proxy} \
-e FTP_PROXY=${FTP_PROXY} -e http_proxy=${http_proxy} \
-e HTTP_PROXY=${HTTP_PROXY} -e https_proxy=${https_proxy} \
-e HTTPS_PROXY=${HTTPS_PROXY} -e no_proxy=${no_proxy} \
-e NO_PROXY=${NO_PROXY} -e socks_proxy=${socks_proxy} \
-e SOCKS_PROXY=${SOCKS_PROXY}"Run the workflow with the docker run command, as shown:
export CONFIG_DIR=$PWD/../configs
export DATASET_DIR=$PWD/../data
export OUTPUT_DIR=$PWD/../output
docker run -a stdout ${DOCKER_RUN_ENVS} \
-v /$PWD/../hf_nlp:/workspace/hf_nlp \
-v /$PWD/../vision_wf:/workspace/vision_wf \
-v /${CONFIG_DIR}:/workspace/configs \
-v /${DATASET_DIR}:/workspace/data \
-v /${OUTPUT_DIR}:/workspace/output \
--privileged --init -it --rm --pull always \
intel/ai-workflows:pa-disease-prediction \
bashRun the command below for fine-tuning and inference:
python src/breast_cancer_prediction.py --config_file /workspace/configs/disease_prediction_baremetal.yamlStop containers created by docker compose and remove them.
docker compose down- Install Helm
curl -fsSL -o get_helm.sh https://raw.githubusercontent.com/helm/helm/main/scripts/get-helm-3 && \
chmod 700 get_helm.sh && \
./get_helm.sh- Install Argo Workflows and Argo CLI
- Configure your Artifact Repository
- Ensure that your dataset and config files are present in your chosen artifact repository.
export NAMESPACE=argo
helm install --namespace ${NAMESPACE} --set proxy=${http_proxy} disease-prediction ./chart
argo submit --from wftmpl/disease-prediction --namespace=${NAMESPACE}To view your workflow progress
argo logs @latest -fUsers are encouraged to use python virtual environments for consistent package management
Using virtualenv:
cd $WORKSPACE/brca_multimodal
python3.9 -m venv hls_env
source hls_env/bin/activate
Or conda: If you don't already have conda installed, see the Conda Linux installation instructions.
cd $WORKSPACE/brca_multimodal
conda create --name hls_env python=3.9
conda activate hls_env
This step involves the installation of the following workflows:
- HF Fine-tune & Inference Optimization workflow
- Transfer Learning based on TLT workflow
bash setup_workflows.sh
To train the multi-model disease prediction, utilize the 'breast_cancer_prediction.py' script along with the arguments outlined in the 'disease_prediction_baremetal.yaml' configuration file, which has the following structure:
%%{init: {'theme': 'dark'}}%%
flowchart LR
disease_prediction_baremetal.yaml --> output_dir
disease_prediction_baremetal.yaml --> write
disease_prediction_baremetal.yaml --> nlp
nlp --> nlp-finetune-params
nlp --> nlp-inference-params
nlp --> other-nlp-params
disease_prediction_baremetal.yaml --> vision
vision --> vision-finetune-params
vision --> vision-inference-params
vision --> other-vision-params
The 'disease_prediction_baremetal.yaml' file includes the following parameters:
-
output_dir: specifies the location of the output model and inference results
-
write: a container parameter that is set to false for bare metal
-
nlp:
- finetune: runs nlp fine-tuning
- inference: runs nlp inference
- additional parameters for the HF fine-tune and inference optimization workflow (more information available here)
-
vision:
- finetune: runs vision fine-tuning
- inference: runs vision inference
- additional parameters for the Vision: Transfer Learning Toolkit based on TLT workflow (more information available here)
To solely perform the fine-tuning process, set the 'finetune' parameter to true in the 'disease_prediction.yaml' file and execute the following command:
python src/breast_cancer_prediction.py --config_file configs/disease_prediction_baremetal.yaml
After the models are trained and saved using the script from step 4, load the NLP and vision models using the inference option. This applies a weighted ensemble method to generate a final prediction. To only run inference, set the 'inference' parameter to true in the 'disease_prediction.yaml' file and run the command provided in step 4.
Alternatively, you can combine the training and inference processes into one execution by setting both the 'finetune' and 'inference' parameters to true in the 'disease_prediction.yaml' file and running the command provided in step 4.
To be able to run the code inside brca_multimodal_notebook.ipynb user should first create a virtual environment. For conda use the following commands:
conda create --name hls_env python=3.9 ipykernel jupyterlab tornado==6.2 ipywidgets==8.0.4 -y -q
To activate it run the following command:
conda activate hls_env
python -m ipykernel install --user --name hls_env
Once Jupyter Lab is installed run from the project root directory inside hls_env environment:
jupyter lab
When jupyter lab is running click on the provided link and follow the instructions inside the notebook. If needed change the kernel to hls_env.
A successful execution of inference returns the confusion matrix of the sub-models and ensembled model, as shown in these example results:
------ Confusion Matrix for Vision model ------
Benign Malignant Normal Precision
Benign 18.0 11.000 1.000 0.486
Malignant 5.0 32.000 0.000 0.615
Normal 14.0 9.000 25.000 0.962
Recall 0.6 0.865 0.521 0.652
------ Confusion Matrix for NLP model ---------
Benign Malignant Normal Precision
Benign 25.000 4.000 1.0 0.893
Malignant 3.000 34.000 0.0 0.895
Normal 0.000 0.000 48.0 0.980
Recall 0.833 0.919 1.0 0.930
------ Confusion Matrix for Ensemble --------
Benign Malignant Normal Precision
Benign 26.000 4.000 0.0 0.897
Malignant 3.000 34.000 0.0 0.895
Normal 0.000 0.000 48.0 1.000
Recall 0.867 0.919 1.0 0.939
This Github repo describes a reference kit for multi-modal disease prediction in the biomedical domain. The kit provides an end-to-end solution for fine-tuning and inference using categorized contrast-enhanced mammography data and radiologists' notes to predict breast cancer diagnosis. The reference kit includes a vision workload that trains an image classifier using CESM images, and a NLP pipeline that trains a document classifier using annotation notes about a patient's symptoms. Both pipelines create predictions for the diagnosis of breast cancer, which are then combined using a weighted ensemble method to create a final prediction. The ultimate goal of the reference kit is to develop and optimize a decision support system that can automatically categorize samples as normal, benign, or malignant, thereby reducing the need for expert involvement.
As a future work, we will use a postprocessing method that will ensemble the different domain knowledge at feature level and fine-tune a model that would increase the accuracy of the prediction, speed up the end-to-end execution time for fine-tuning and inference, and reduce the cost of computation.
- If you want to enable distributed training on k8s for your use case, please follow steps to apply that configuration mentioned in the Intel® Transfer Learning Tools which provides insights into k8s operators and yml file creation.
Tunable configurations and parameters are exposed using yaml config files allowing users to change model training hyperparameters, datatypes, paths, and dataset settings without having to modify or search through the code.
To deploy this reference use case on a different or customized dataset, you can easily modify the disease_prediction_baremetal.yaml file. For instance, if you have a new text dataset, simply update the paths of finetune_input and inference_input and adjust the dataset features in the disease_prediction_baremetal.yaml file, as demonstrated below.
%%{init: {'theme': 'dark'}}%%
flowchart LR
nlp --> args
args --> local_dataset
local_dataset --> |../data/annotation/training.csv| finetune_input
local_dataset --> |../data/annotation/testing.csv| inference_input
local_dataset --> features
features --> |label| class_label
features --> |symptoms| data_column
features --> |Patient_ID| id
local_dataset --> |Benign, Malignant, Normal| label_list
To implement this reference use case on a different or customized pre-training model, modifications to the disease_prediction_baremetal.yaml file are straightforward. For instance, to use an alternate model, one can update the path of the model by modifying the 'model_name_or_path' and 'tokenizer_name' fields in the disease_prediction_baremetal.yaml file structure. The following example illustrates this process:
%%{init: {'theme': 'dark'}}%%
flowchart LR
nlp --> args
args --> |emilyalsentzer/Bio_ClinicalBERT| model_name_or_path
args --> |emilyalsentzer/Bio_ClinicalBERT| tokenizer_name
For more information or to read about other relevant workflow examples, see these guides and software resources:
- Intel® AI Analytics Toolkit (AI Kit)
- Intel® Neural Compressor
- Intel® Extension for PyTorch
- Intel® Transfer Learning Tool
- Intel® Extension for Transformers
Currently mixing workflows on bare-metal and Docker from same path on the host is not supported. Please start each workflow run from scratch to minimize the chances of previously cached data.
The end-to-end multi-modal disease prediction tea tracks both bugs and enhancement requests using disease prediction GitHub repo. We welcome input, however, before filing a request, search the GitHub issue database.
*Other names and brands may be claimed as the property of others. Trademarks.
Disclaimer: This reference implementation shows how to train a model to examine and evaluate a diagnostic theory and the associated performance of Intel technology solutions using very limited, non-diverse datasets to train the model. The model was not developed with any intention of clinical deployment and therefore lacks the requisite breadth and depth of quality information in its underlying datasets, or the scientific rigor necessary to be considered for use in actual diagnostic applications. Accordingly, while the model may serve as a foundation for additional research and development of more robust models, Intel expressly recommends and requests that this model not be used in clinical implementations or as a diagnostic tool.