The ged-cost-learn-framework project is the code for the paper
Bridging Distinct Spaces in Graph-based Machine Learning (preprint) published in the proceedings of ACPR 2023.
python3 -m pip install graphkit-learn
python3 -m pip install seaborn
# The followings are required to run the GNN models:
python3 -m pip install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118
python3 -m pip install torch-geometric pyg_lib torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.0.0+cu118.html
NVIDIA GPU and driver is required to run the GNN models. Check this tutorial for more information.
git clone [email protected]:jajupmochi/ged-cost-learn-framework.git
cd ged-cost-learning/
python setup.py [develop] [--user]
python3 gcl_frame/models/run_xps.py
The outputs will be in the gcl_frame/models/outputs/ directory.
Align GEDs in graph space G and distances in an embedding space E:
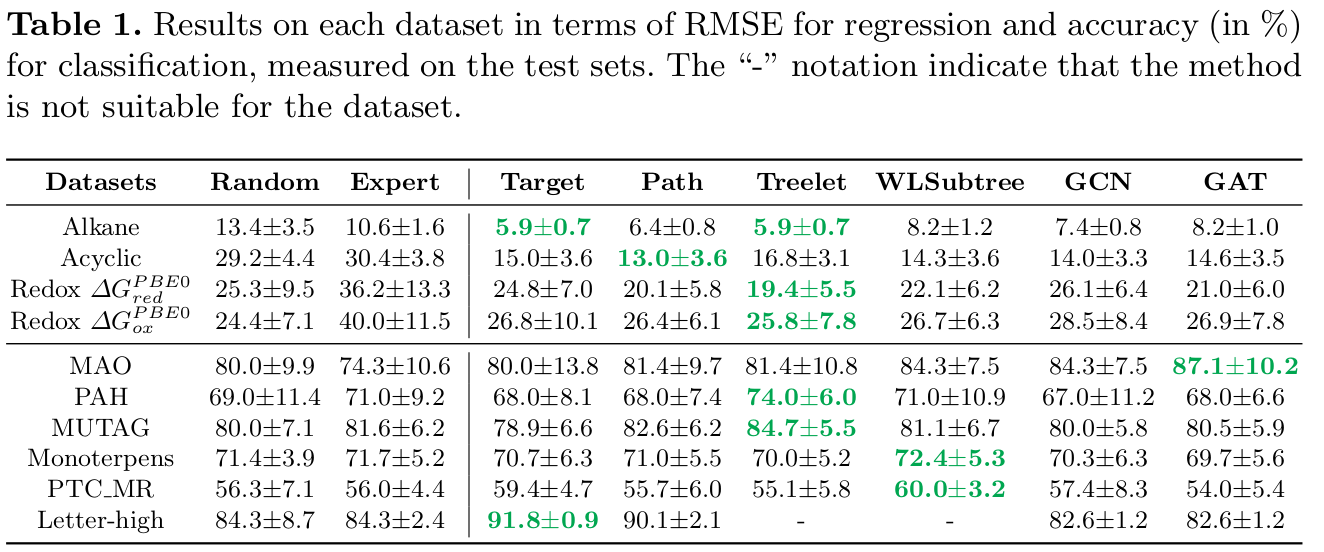
Results on each dataset in terms of RMSE for regression and accuracy (in %) for classification, measured on the test sets. The "-" notation indicate that the method is not suitable for the dataset:
Pre-images constructed by different algorithms for Letter-high with shortest path (SP) and structural shortest path (SSP) kernels:
- Linlin Jia, the PRG group, Institute of Computer Science, University of Bern
- Xiao Ning, State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University
- Benoit Gaüzère, the LITIS Lab, INSA Rouen Normandie
- Paul Honeine, the LITIS Lab, Université de Rouen Normandie
- Kaspar Riesen, the PRG group, Institute of Computer Science, University of Bern
If you have used this library in your publication, please cite the following paper:
@inproceedings{jia2023bridging,
title={Bridging Distinct Spaces in Graph-Based Machine Learning},
author={Jia, Linlin and Ning, Xiao and Ga{\"u}z{\`e}re, Benoit and Honeine, Paul and Riesen, Kaspar},
booktitle={Asian Conference on Pattern Recognition},
pages={1--14},
year={2023},
organization={Springer}
}
This research was supported by Swiss National Science Foundation (SNSF) Project No. 200021_188496. The authors would like to thank the COBRA lab (Chimie Organique Bioorganique: Réactivité et Analyse) and the ITODYS lab (Le laboratoire Interfaces Traitements Organisation et DYnamique des Systèmes) for providing the Redox dataset.