This repository was forked from the excellent tutorial by Chris Fonnesbeck: PyMC3 DataScienceLA.
CEAi has modified some material and added new material - especially exercises with the aim of using this series of notebooks to prepare for our first Precision workshop - on Bayesian modelling.
At CEAi we are always looking to push our ML skills to the next level and this is an experiment in how to implement this in the fast paced environment of a startup studio. Instead of just doing a workshop on Bayesian modelling, we believe that we can get much more out of a workshop if we invest significant effort into preparation. We are working through this material in the course of approximately 3 months to prepare for a workshop that will take approximately 3 days. We expect at least the following effects: everyone will enter the workshop with a given minimal level of knowledge, the three month period will allow lots of time to get into the topics and think through more difficult issues, we will have basic coding experience with PyMC3 as a vehicle for learning about Bayesian modelling and since these materials are being created just before use, we can adjust the difficulty level online.
We welcome all feedback to the notebooks and please send us pull requests with ideas for improvement.
We also note that we have found lots of inspiration elsewhere for arranging this material, including the PyMC3 documentation, examples, various talks and papers. Relevant sources should be appended to each notebook.
- Measure initial skill level and knowledge state of group
- Work out speed and method of proceeding at group level
- Come up with a time plan targeting May for completion (deadline Apr 15th)
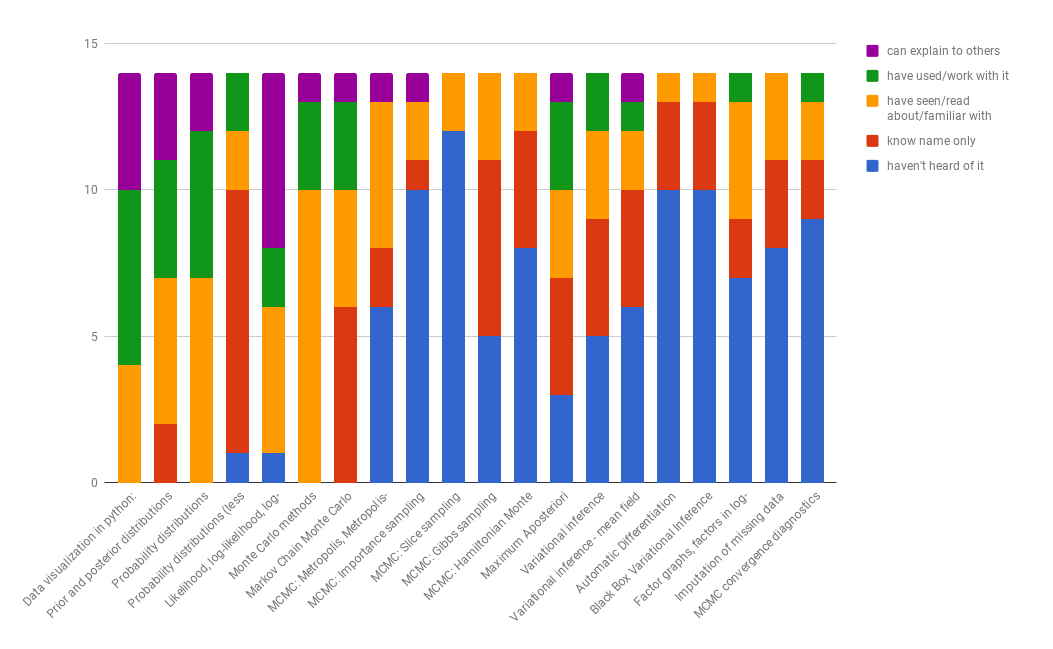
We polled the ML group in CEAi as to the actual state of knowledge (self-reported) of various topics stated below and the following stacked graph displays the result.
We aim for another measurement on 22.3.2018.
We order the topics as follows: building models (using PyMC3 for inference), inference based on sampling, inference based on variational methods and finally model checking. We aim for different levels of depth depending on the topic based on estimating what can be best handled in-house and what is best left for the workshop itself.
- Variable types
- Probability models
- Well known distributions refresher
- Simple case studies
- Comparing analytical solutions to numeric approximations
- Beta-Binomial model
- Normal-Normal model with known precision
- Specifying priors and likelihoods
- Deterministic variables
- Factor potentials
- Custom variables
- Case study: fitting a 9x9 pixel faces
- Case study: tagging two-token names
- Monte Carlo: importance sampling and rejection sampling
- Markov Chain Monte Carlo
- Metropolis
- Examples: 2D Gaussian, linear regression with Laplace prior
- Complex study: Responses to visual stimuli study
- Exercise: write your own sampler
- Gibbs sampling and Metropolis-within-Gibbs
- Example: Ising model
- Example: Metropolis within Gibbs toy example
- Example: Response times to visual stimuli Metropolis-within-Gibbs
- Hamiltonian Monte Carlo
- Convergence diagnostics
- Goodness of fit
- Plotting and summarization
Running PyMC3 requires a working Python interpreter, either version 2.7 (or more recent) or 3.4 (or more recent); we recommend that new users install version 3.5 (but see special note below if you are a Windows user). A complete Python installation for Mac OSX, Linux and Windows can most easily be obtained by downloading and installing the free Anaconda Python Distribution by ContinuumIO.
PyMC3 can be installed using conda, a package management tool that is bundled with Anaconda. PyMC3 also depends on several third-party Python packages which will be automatically installed when installing via conda. The four required dependencies are: Theano, NumPy, SciPy, Matplotlib, and joblib. To take full advantage of PyMC3, the optional dependencies seaborn, pandas and Patsy should also be installed. You can install PyMC3 and its dependencies by cloning this repository:
git clone https://github.com/oapio/PrecisionWorkshop1_Prep
Then move into the directory created by the clone, and install the required packages using conda:
cd PyMC3_DataScienceLA
conda env create -f environment.ymlThis will create a virtual environment called pymc_tutorial that includes the dependencies for PyMC3 that is completely separate from any other Python installations you may have on your machine. To activate this environment to run the course materials, you can run the following command from the terminal:
source activate pymc_tutorialIf you would rather not install the software yourself, you can use the MyBinder.org link at the top of the page to run the course materials on a remote server