This package is the R code for the FAst DIstributed
(FADI) PCA method for federated data when both the dimension
Our paper is here:
Shen, S., Lu, J. and Lin, X., 2023. FADI: Fast Distributed Principal Component Analysis With High Accuracy for Large-Scale Federated Data. arXiv preprint arXiv:2306.06857.
The R codes folder contains the R scripts for simulation studies, and application of FADI to the 1000 Genomes data (estimation of principal eigenspace and inferential analysis under the degree-corrected mixed membership model).
R scripts example_spiked_covariance.R, example_GMM.R, example_DCMM.R, and example_missing_matrix.R contain the simulation codes for implementing FADI under the spiked covariance model, the Gaussian mixture models (GMM), the degree-corrected mixed membership (DCMM) model, and the incomplete matrix inference model respectively. Input parameters are d-dimension of data, mc-index of independent Monte Carlo simulations, and rt-ratio of
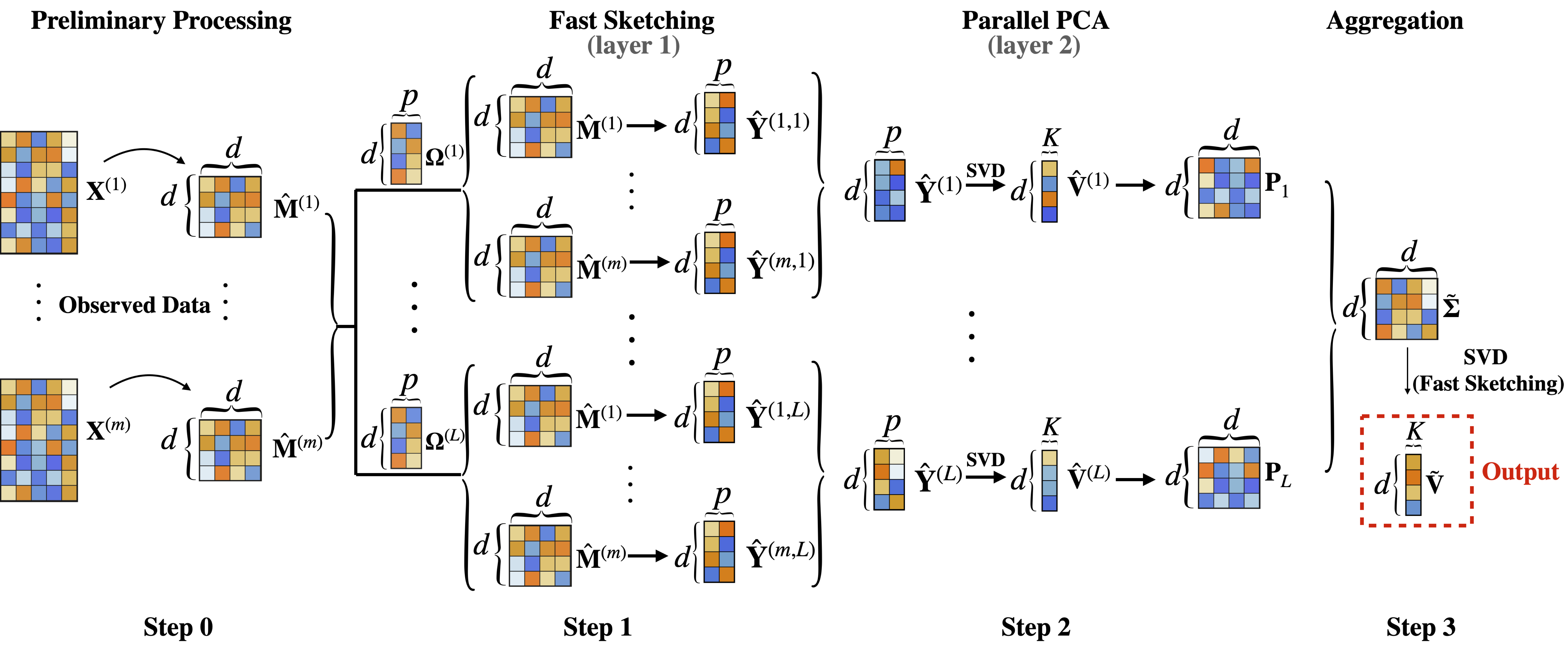
R scripts 1000g_estimation_layer_1.R and 1000g_estimation_layer_2.R contain the codes for applying FADI for estimating the principal eigenspace of the 1000 Genomes data. 1000g_estimation_layer_1.R implements Step 1 of FADI, with input parameters i-index for distributed data split, l-index for parallel sketching, and p-dimension of fast sketching. 1000g_estimation_layer_2.R implements step 2 of FADI, with input parameters l-index for parallel sketching, and p-dimension of fast sketching.
R script inference_1000g_SBM.R implements Step 1 and Step 2 of FADI for computing the top PCs of the undirected graph generated based on the 1000 Genomes data, with input parameter l-index for parallel sketching. R script multiple_testing_1000g.Rmd performs multiple testing on inferring subject population of the 1000 Genomes data. The script multiple_testing_1000g.Rmd first implements Step 3 of FADI, by aggregating the parallel sketching results to output the FADI estimator of the top PCs. Then the script multiple_testing_1000g.Rmd performs the inferential procedure for membership testing using the FADI PC estimator, as detailed in Supplement D of the paper "Dimension Reduction for Large-Scale Federated Data: Statistical Rate and Asymptotic Inference".
The folder Data contains supplementary data for applying FADI to inferential analysis of the 1000 Genomes data. The file 1000g_sbm95.RData contains an undirected graph generated based on the 1000 Genomes data, used for the inferential application of FADI, and the file 1KG_TRACE_pca.txt contains the population information of the 1000 Genomes data subjects.