A paper introducing jMetalPy is available at: https://doi.org/10.1016/j.swevo.2019.100598
You can install the latest version of jMetalPy with pip,
pip install jmetalpy # or "jmetalpy[distributed]"Notes on installing with pip
jMetalPy includes features for parallel and distributed computing based on pySpark and Dask.
These (extra) dependencies are not automatically installed when running pip, which only comprises the core functionality of the framework (enough for most users):
pip install jmetalpyThis is the equivalent of running:
pip install "jmetalpy[core]"Other supported commands are listed next:
pip install "jmetalpy[dev]" # Install requirements for development
pip install "jmetalpy[distributed]" # Install requirements for parallel/distributed computing
pip install "jmetalpy[complete]" # Install all requirementsExamples of configuring and running all the included algorithms are located in the documentation.
from jmetal.algorithm.multiobjective import NSGAII
from jmetal.operator import SBXCrossover, PolynomialMutation
from jmetal.problem import ZDT1
from jmetal.util.termination_criterion import StoppingByEvaluations
problem = ZDT1()
algorithm = NSGAII(
problem=problem,
population_size=100,
offspring_population_size=100,
mutation=PolynomialMutation(probability=1.0 / problem.number_of_variables, distribution_index=20),
crossover=SBXCrossover(probability=1.0, distribution_index=20),
termination_criterion=StoppingByEvaluations(max_evaluations=25000)
)
algorithm.run()We can then proceed to explore the results:
from jmetal.util.solution import get_non_dominated_solutions, print_function_values_to_file,
print_variables_to_file
front = get_non_dominated_solutions(algorithm.result())
# save to files
print_function_values_to_file(front, 'FUN.NSGAII.ZDT1')
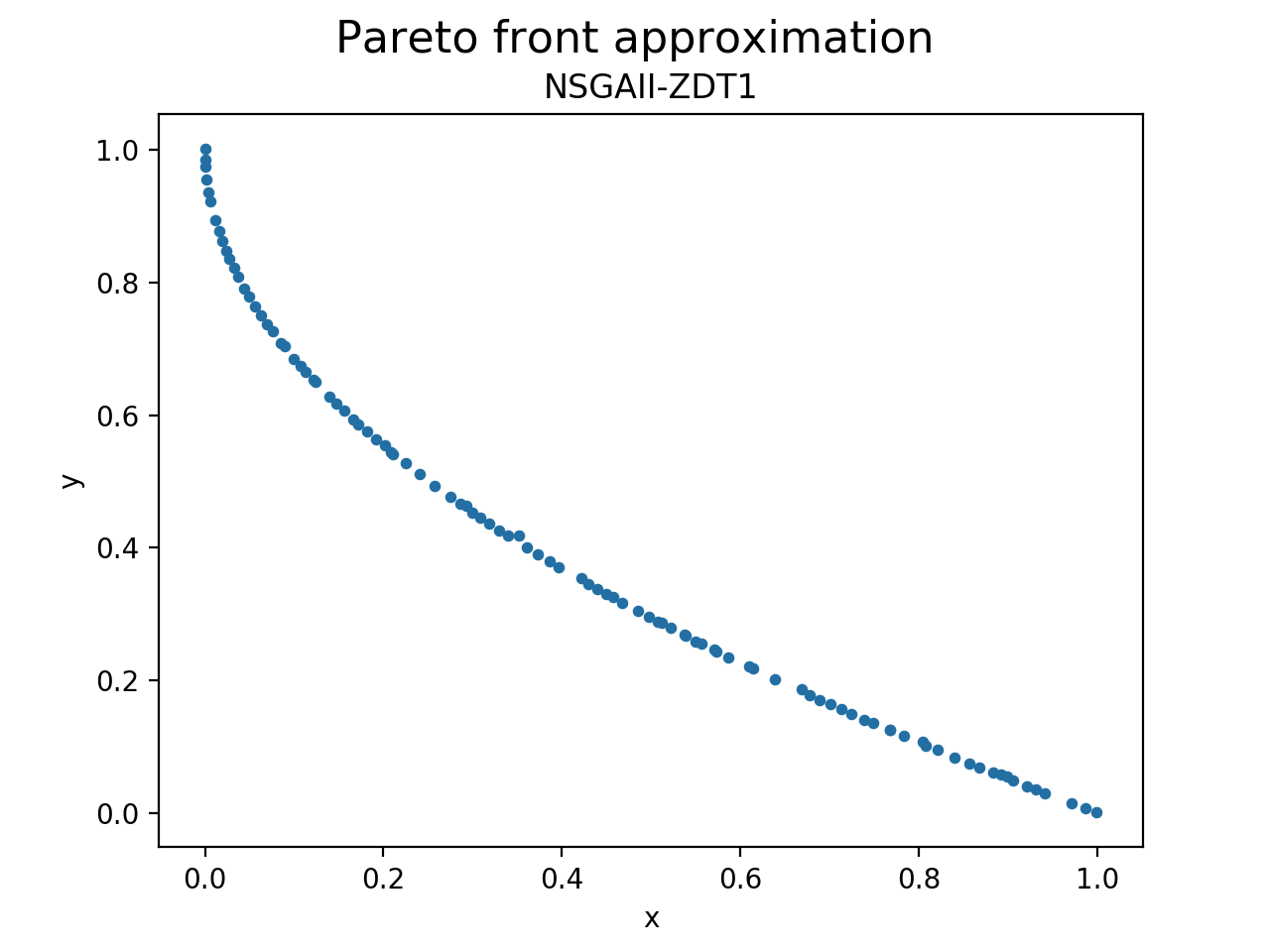
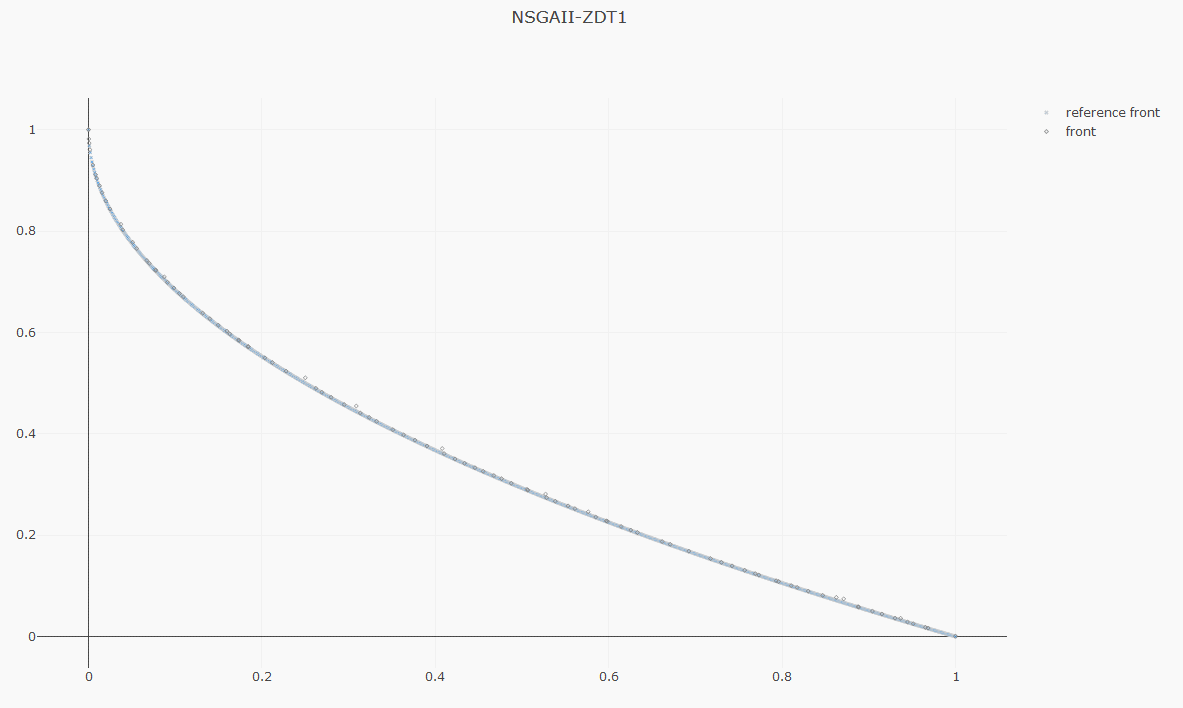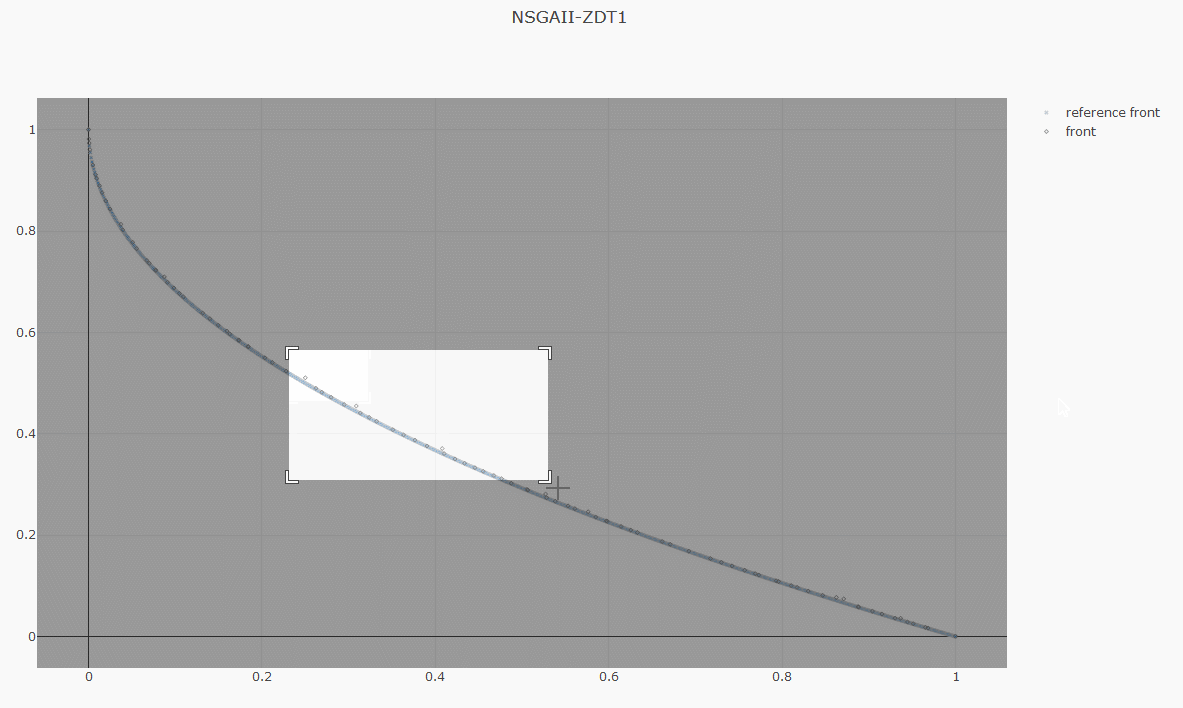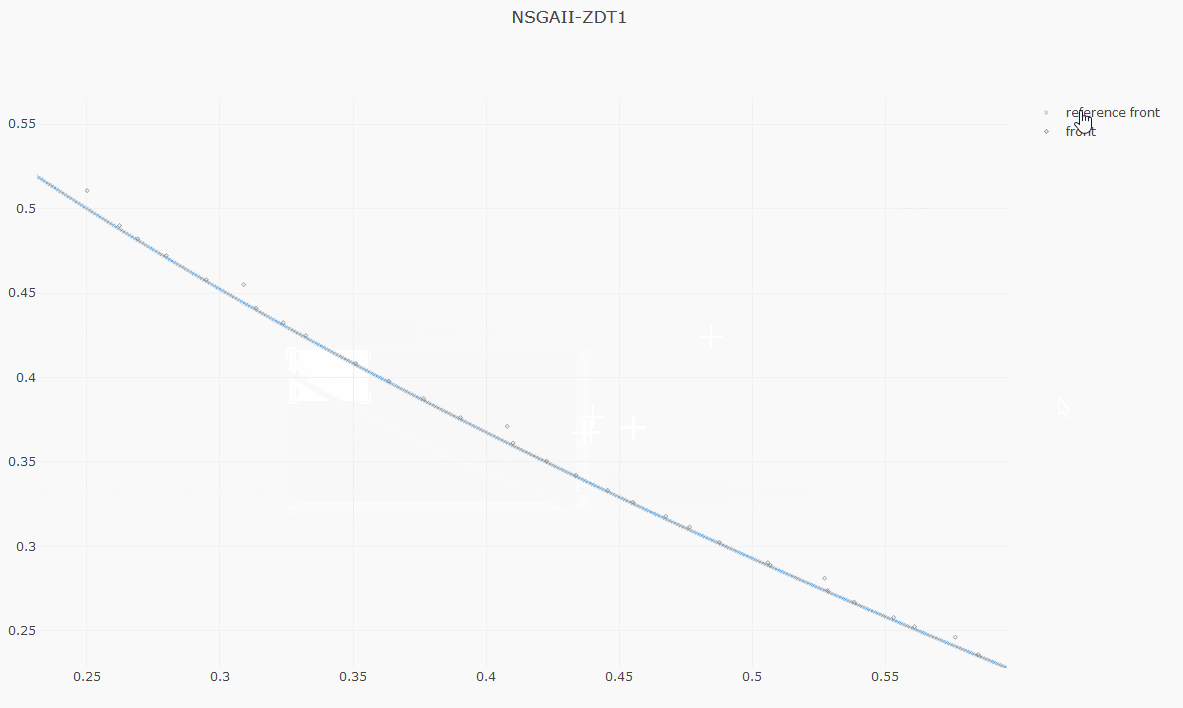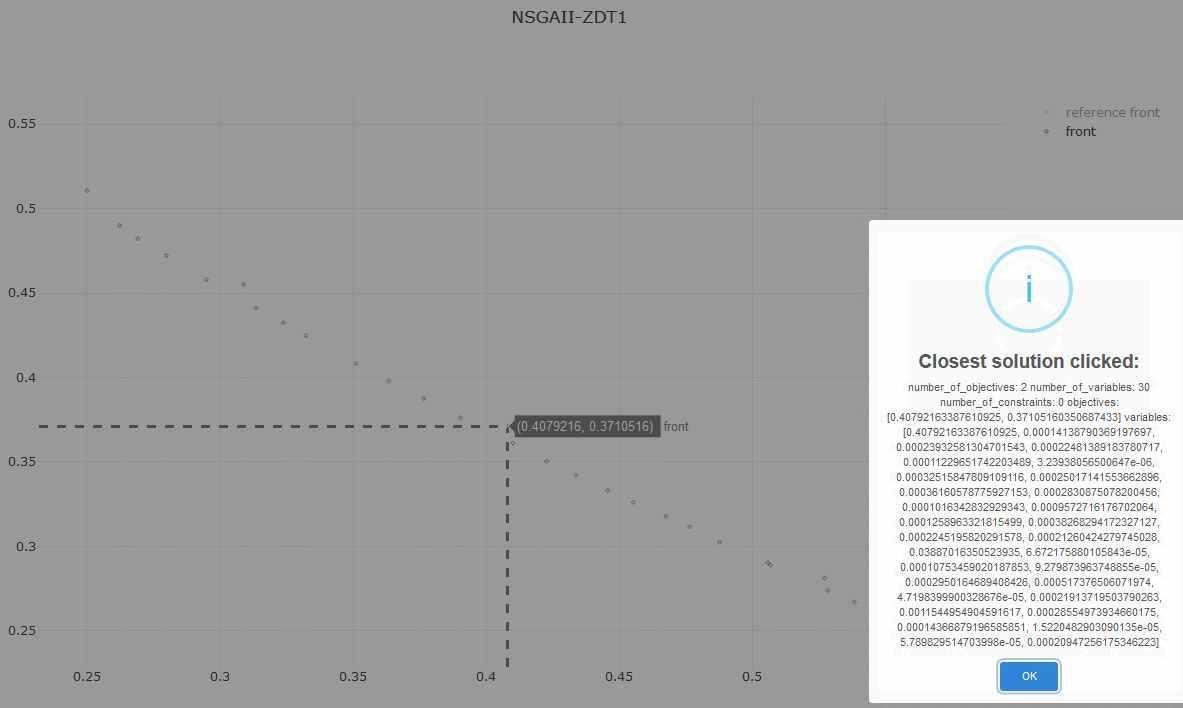
print_variables_to_file(front, 'VAR.NSGAII.ZDT1')Or visualize the Pareto front approximation produced by the algorithm:
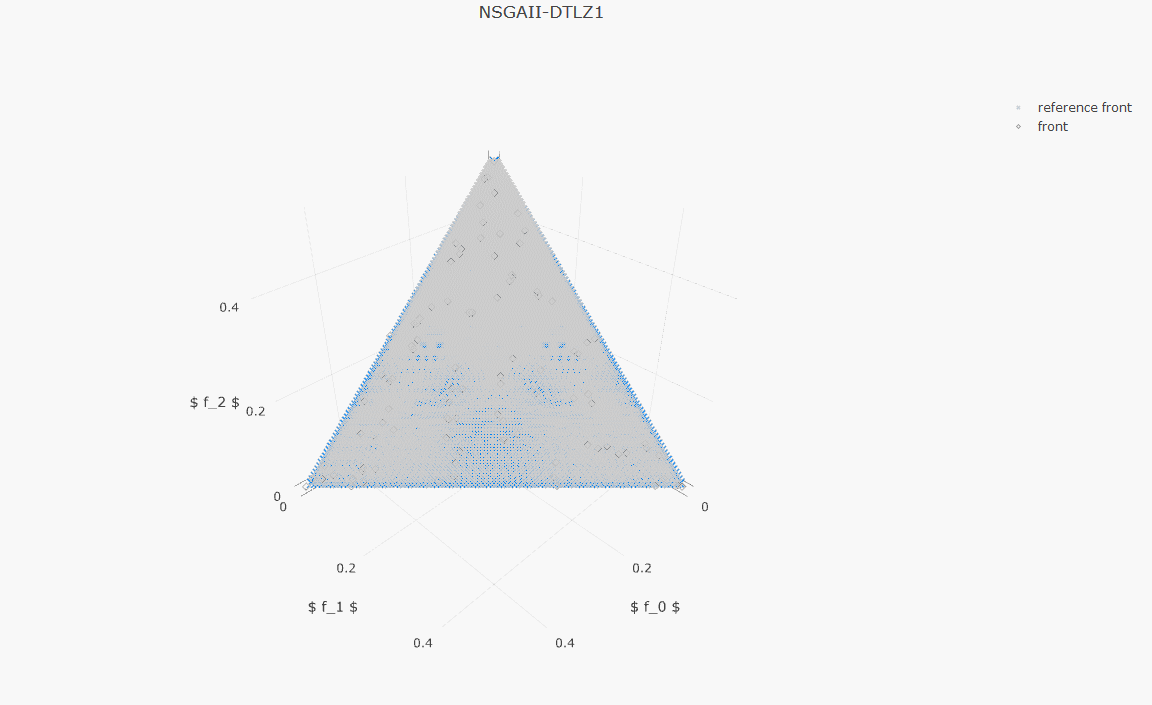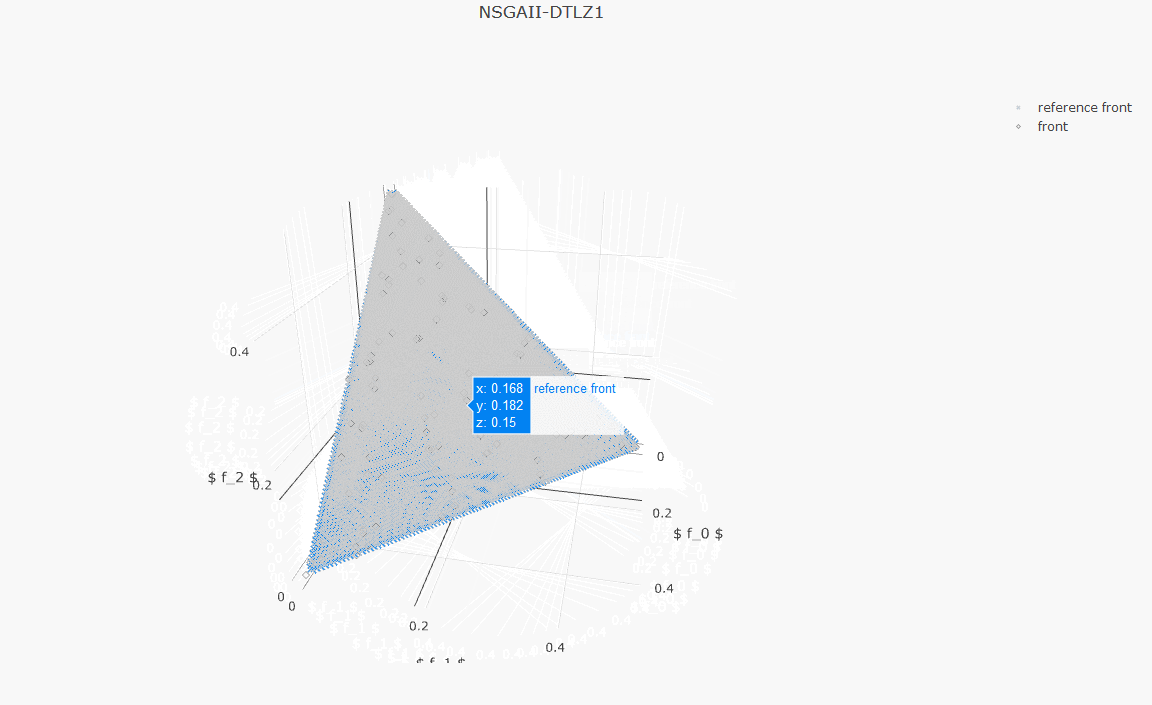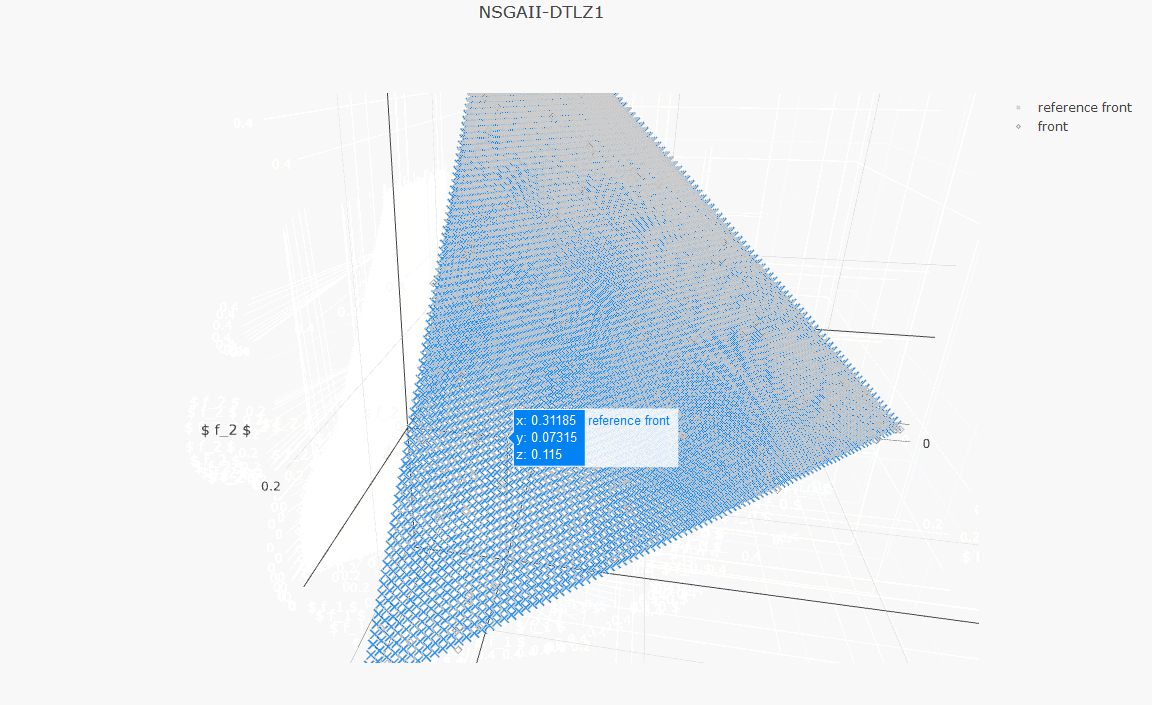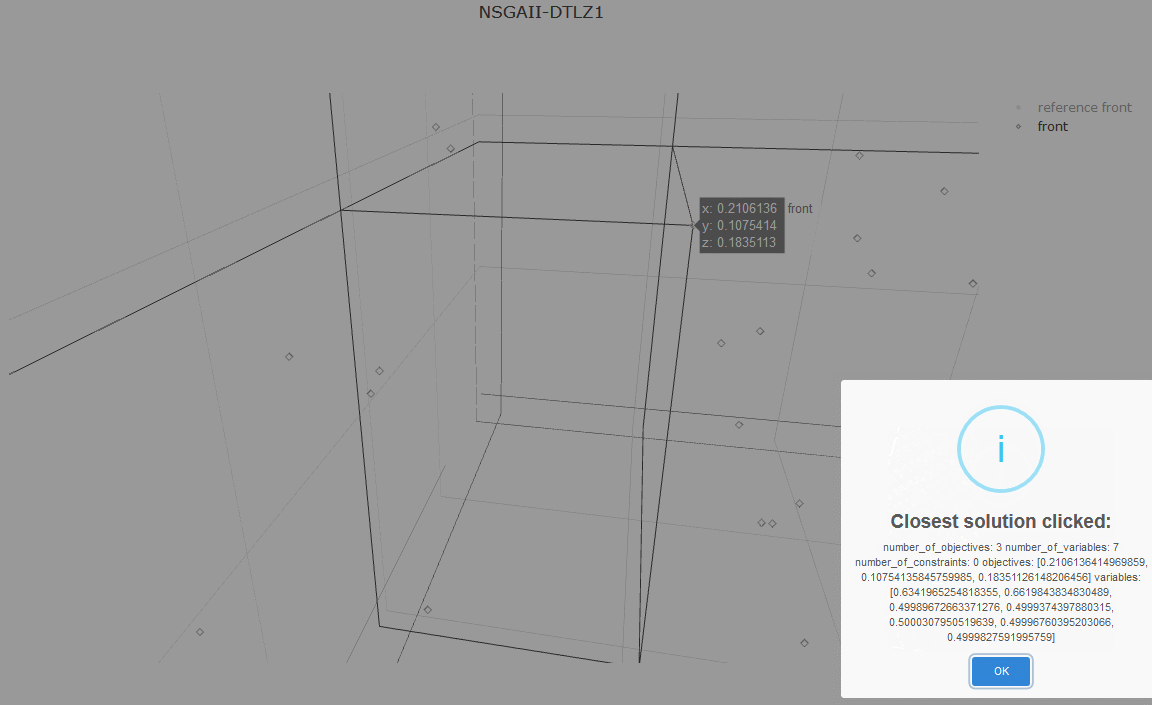
from jmetal.lab.visualization import Plot
plot_front = Plot(title='Pareto front approximation', axis_labels=['x', 'y'])
plot_front.plot(front, label='NSGAII-ZDT1', filename='NSGAII-ZDT1', format='png')The current release of jMetalPy (v1.7.0) contains the following components:
- Algorithms: local search, genetic algorithm, evolution strategy, simulated annealing, random search, NSGA-II, NSGA-III, SMPSO, OMOPSO, MOEA/D, MOEA/D-DRA, MOEA/D-IEpsilon, GDE3, SPEA2, HYPE, IBEA. Preference articulation-based algorithms (G-NSGA-II, G-GDE3, G-SPEA2, SMPSO/RP); Dynamic versions of NSGA-II, SMPSO, and GDE3.
- Parallel computing based on Apache Spark and Dask.
- Benchmark problems: ZDT1-6, DTLZ1-2, FDA, LZ09, LIR-CMOP, RWA, unconstrained (Kursawe, Fonseca, Schaffer, Viennet2), constrained (Srinivas, Tanaka).
- Encodings: real, binary, permutations.
- Operators: selection (binary tournament, ranking and crowding distance, random, nary random, best solution), crossover (single-point, SBX), mutation (bit-blip, polynomial, uniform, random).
- Quality indicators: hypervolume, additive epsilon, GD, IGD.
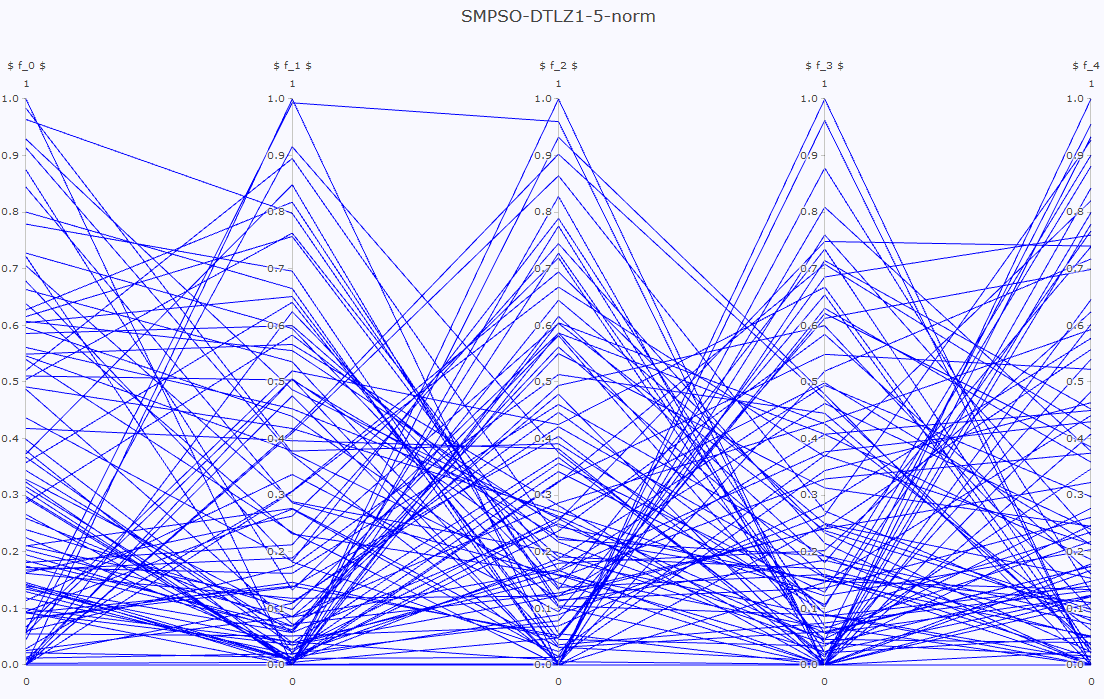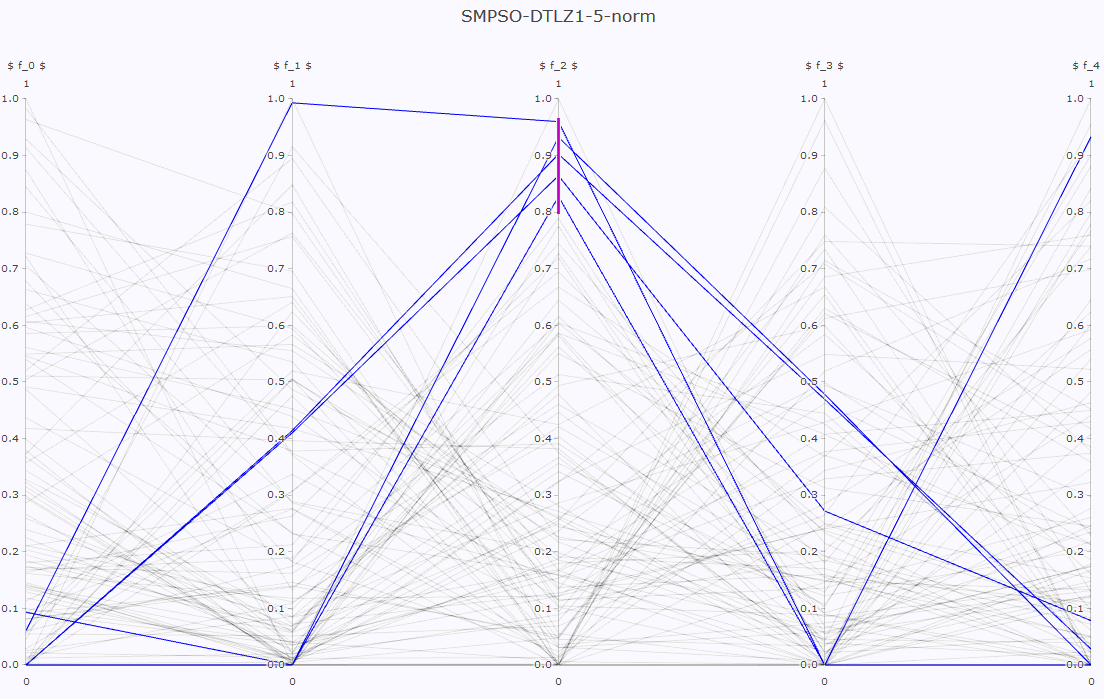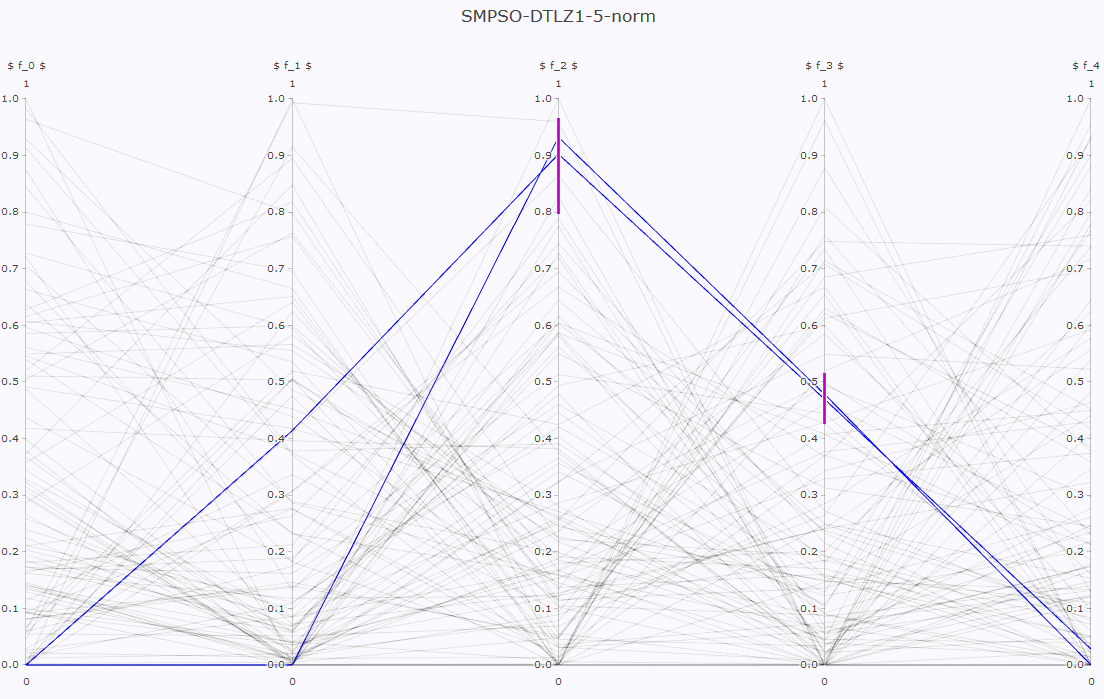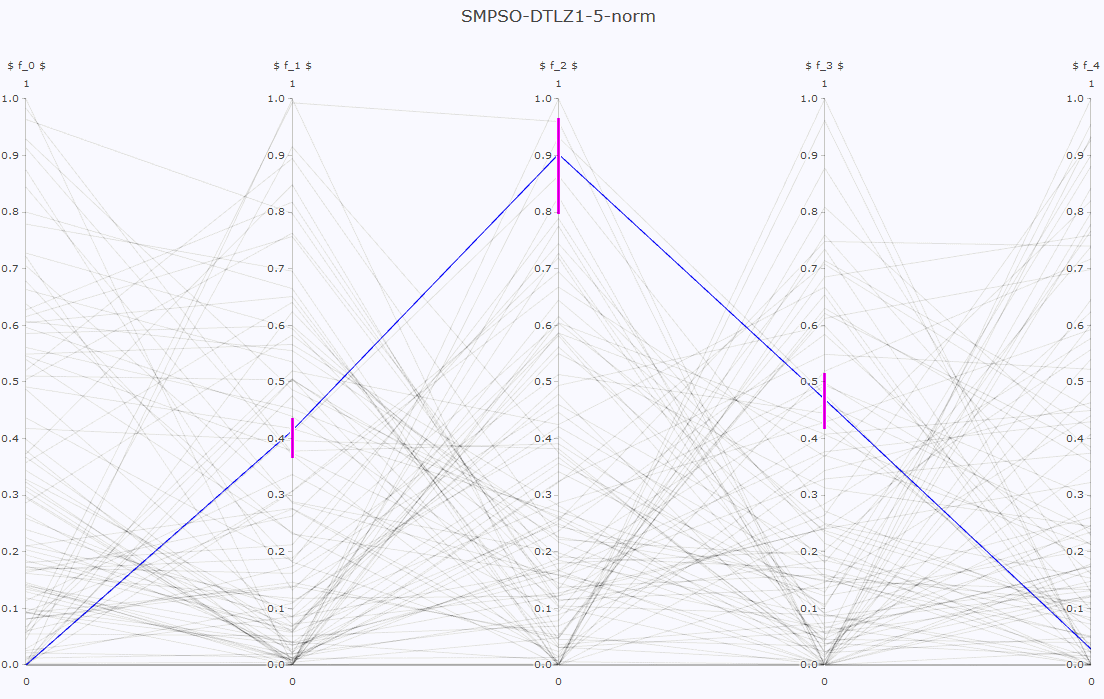
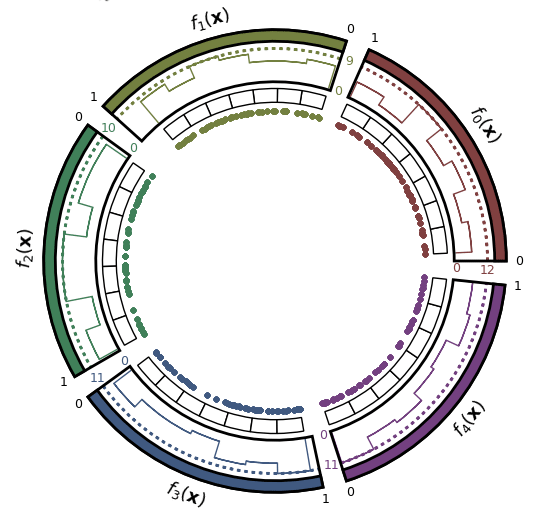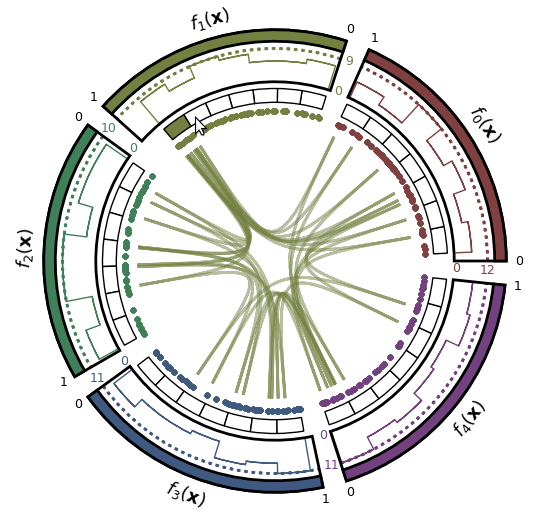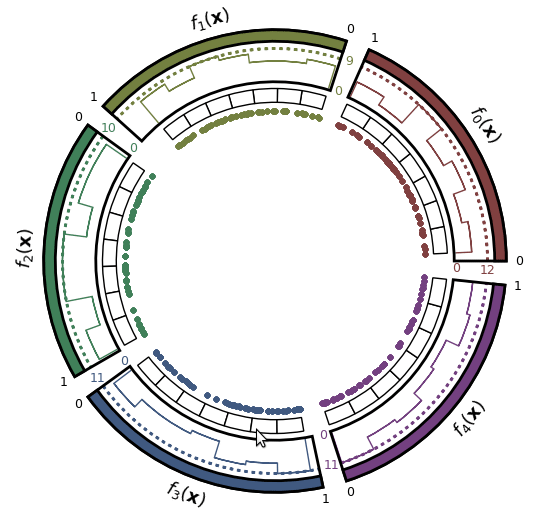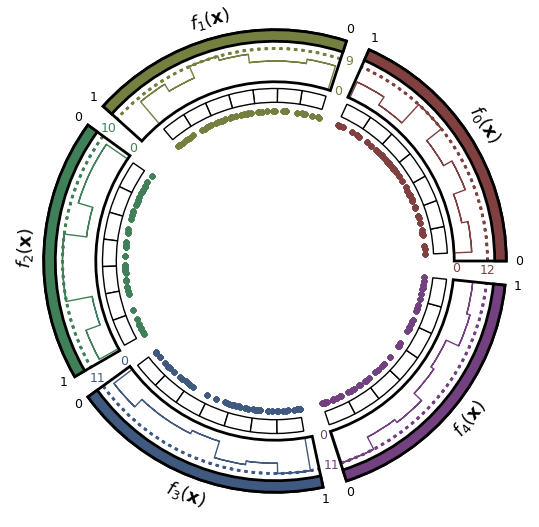
- Pareto front approximation plotting in real-time, static or interactive.
- Experiment class for performing studies either alone or alongside jMetal.
- Pairwise and multiple hypothesis testing for statistical analysis, including several frequentist and Bayesian testing methods, critical distance plots and posterior diagrams.
 |
 |
|---|---|
 |
 |
- [v1.7.0] Add RWA benchmark, refactor classes BinarySolution and BinaryProblem.
- [v1.6.0] Refactor class Problem, the single-objective genetic algorithm can solve constrained problems, performance improvements in NSGA-II, generation of Latex tables summarizing the results of the Wilcoxon rank sum test, added a notebook folder with examples.
- [v1.5.7] Use of linters for catching errors and formatters to fix style, minor bug fixes.
- [v1.5.6] Removed warnings when using Python 3.8.
- [v1.5.5] Minor bug fixes.
- [v1.5.4] Refactored quality indicators to accept numpy array as input parameter.
- [v1.5.4] Added CompositeSolution class to support mixed combinatorial problems. #69
This project is licensed under the terms of the MIT - see the LICENSE file for details.