- simulate an ABT having stochastic output distribution for each leaf.
- Use hmmlearn library to estimate stochastic ABT parameters from output data.
- Support authoring of ABTs.
- ABTs and related Hidden Markov Models are described in "Hidden Markov Models derived from Behavior Trees"
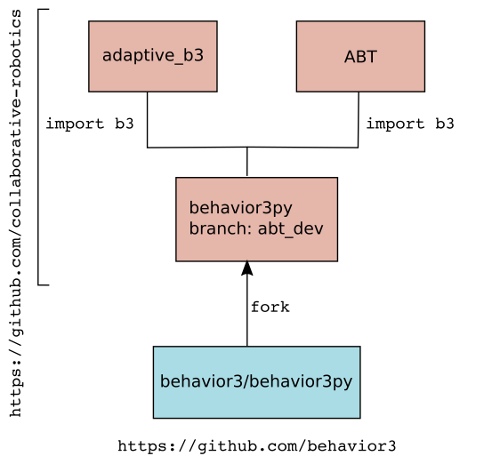
The simulation is built on the b3 library (a fork of
behavior3py, as modified in the abt_dev branch
a python Behavior Tree library. The new leaf class, aug_leaf(b3.Action) inherits the
b3 Action class and adds transition and emission probabilities.
Emmisions (observations) from each state are modeled by NSYMBOLS discrete symbols.
There is a discrete Gaussian probability distribution applied to each state with mean mu and standard deviation sigma. The observations are integer valued. Default sigma = 2.00.
"Perturbuation" refers to the process of changing the HMM parameters between generating model outputs and running HMM-based analyses (e.g. Forward/Viterbi/Baum-Welch). This is more realistic than starting out with exact parameters.
To build a model you have to create three files (bad!)
-
modelxx.py. This file contains/initializes-
The names of your ABT leaves,
-
The initial probability of success for each leaf
-
The mean and variance for the emission symbol distribution for each state
-
all of this is folded into an object of class model() (defined in abtclass.py)
-
-
[ProjName]_ABT.py. This file sets up the ABT with-
the new leaf class
aug_leaf()(from abtclass.py) -
the 16-state file peg_ABT.py an example peg-in-hole task.
** or-** a simple test project:
simp_ABT.pywith 4+2=6 states** or-** customize your own
Nstate model.- ToDo: automatic generation of HMM from BT file
-
-
top_level_hmm.py. This top-level file seqences the computational steps, typically:-
model setup
-
simulated data generation
-
HMM initialization
-
HMM model fitting
-
reporting
-
Run tl_bw_hmm.py (or whatever you want to call it) with two command line arguments: the HMM perturbation magnitude (0.0<x<0.5) and a text comment for the metadata file.
Your results will appear in three files
-
metadata.txt-- one line for each run of your top-level script. Unique filenames are generated using URI's. Each line contains- date and time stamp
- name of data file
- ownname (name of the top level file)
- git hash (1st 10 chars of current git hash) (for maximum provenance, be sure to commit prior to running your code).
- number of HMM / BT states
- text field (comment)
-
sequences/contains the simulated state transition/observation sequences -
bw_output/containts datafiles (item 2 of metadata above). Data file line:- Task code: (2=Baum Welch)
- Ratio: (codeword mean spacing / sigma)
- di: (codeword spacing)
- HMM_delta: amt HMM params changed
- Sigma: standard deviation of output observations in a state
- run#
- e2 (RMS error between original A matrix and identified A matrix)
- emax (max error between original A matrix and identified A matrix )
-
hmm_bt.pyUtilities for A-matrices, and special magic for setting up an HMM_setup
-
abtclass.pyThe new "leaf" class which Augments the BT to work with Success/Fail probabilities, and stochastic observations from each leaf.
- Now unified with the python
unittestlibrary - To run all tests:
> python2 -m unittest discover tests/
-
Tests now include a random.seed(integer) initialization for deterministic results. Both random and np.random seeds are initialized. (note that there is still some variability though small. Maybe hmmlearn or other module deps rely on a third random number package. Also, some tests run differently when run as above vs individually as below!)
-
tests/test_seq_stats.py runs "correctly" as an individual test (below), but fails as part of the
unittest discovercommand (above). Not clear why yet. -
To run tests individually
> python2 -m unittest tests.XX_TEST_NAMEXX
Important: use . instead of / and DO NOT add the .py extension to test name (complain to unittest developers etc).
Behavior3 library, originally written in Javascript and converted to Python2.7
-
Based on the work of (Marzinotto et al., 2014), in which they propose a formal, consistent and general definition of Behavior Trees;