| Colab | Recep Oğuz Keser |
|---|
Accurate classification of EEG signals for motor and imagined movements is essential for the development of brain-computer interfaces (BCIs) that benefit impaired individuals and enhance functionalities for healthy users. This study explores various classification methods to improve the accuracy and interpretability of EEG signals. Utilizing a dataset of over 1500 EEG recordings from 109 volunteers, subjects performed a series of motor and imagery tasks while their EEGs were recorded using a 64-channel setup and sampled at 160 Hz[7]. The data underwent preprocessing, including filtering and epoch segmentation, to prepare it for analysis. The core of our model is the EEGNet architecture, designed to capture spatial and temporal features through convolutional layers, depthwise separable convolutions, and robust regularization techniques[8][9]. The model is trained using StratifiedKFold cross-validation, with performance metrics such as accuracy, sensitivity, specificity, PPV, and NPV being evaluated.
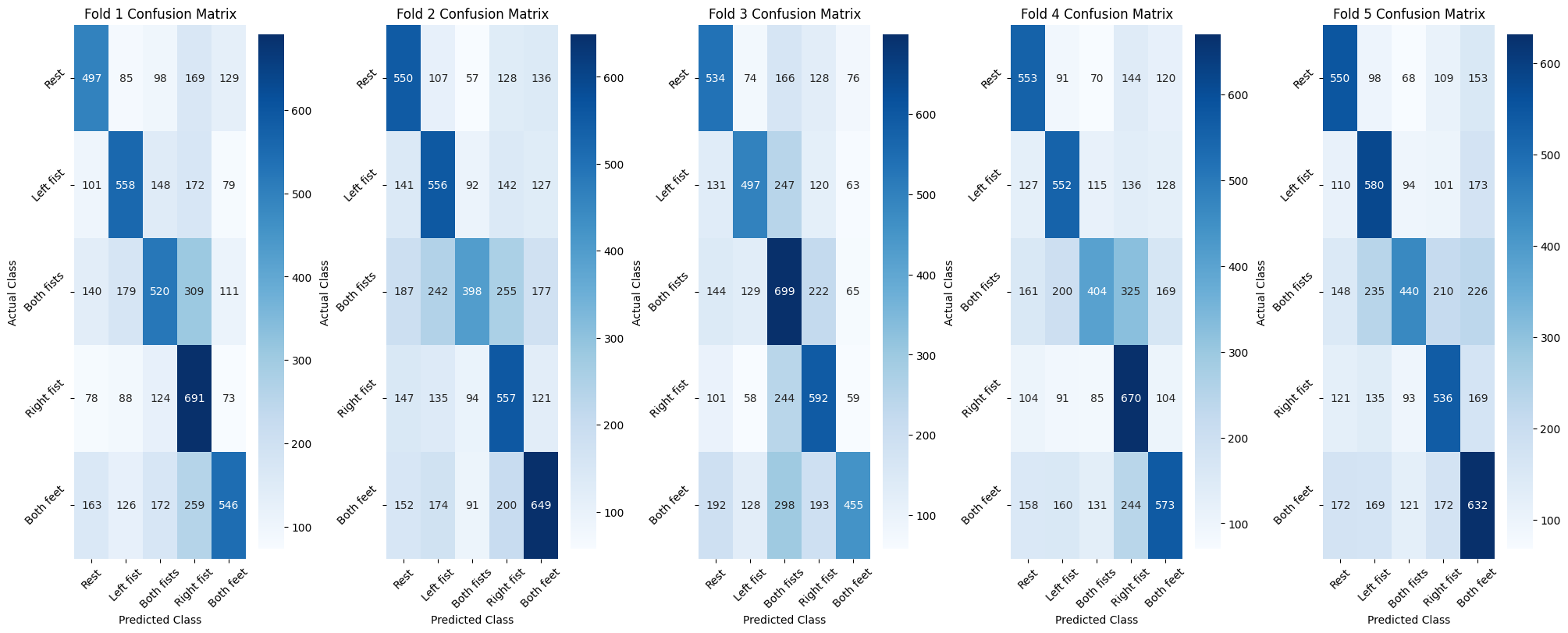
Our results indicate a varying performance across different folds, with significant challenges in accurately classifying the "Both fists" and "Rest" classes. The model achieves high overall accuracy but demonstrates a need for further refinement to consistently distinguish between similar classes. This study highlights the potential of the EEGNet architecture in EEG signal classification while also identifying areas for improvement, such as enhancing the model's ability to differentiate between motor and imagined tasks. Future work could involve separating and independently analyzing motor and imagined tasks to provide deeper insights into their classification challenges and comparing the classification performance across motor, imagined, and combined tasks.
The accurate identification of EEG signals, especially for motor and imagined movements, is essential for advancing brain-computer interfaces (BCIs) that can aid impaired individuals and offer new functionalities for healthy users. Several studies have explored different methods to classify EEG signals with a focus on improving accuracy and interpretability.
Velasco et al. (2023) proposed a multivariate time series approach to enhance the classification accuracy of motor imagery EEG signals, achieving high accuracy with reduced variables, which is essential for avoiding overfitting and improving interpretability[1]. Batistic et al. (2023) explored different classifiers for motor imagery, highlighting the superior performance of a ResNet-based CNN, especially with vibrotactile guidance, suggesting improvements in preprocessing and classifier selection for better accuracy[2]. Alizadeh et al. (2023) utilized a deep CNN to classify motor imagery tasks, showing that 2D wavelet-transformed images and CNN architectures like AlexNet and LeNet provide high classification accuracy, emphasizing the potential of image-based EEG classification approaches[3]. Duan (2023) demonstrated that real and imagined knee movements can be classified with high accuracy using a modified LeNet-5 CNN, suggesting that combining motor imagery and movement execution data could enhance BCI applications[4]. Pham et al. (2023) compared tEEG and conventional EEG, finding that tEEG provides better spatial resolution and signal-to-noise ratio, and when used with CNNs, offers high classification accuracy for finger movements[5].
The dataset used in this project comprises over 1500 EEG recordings from 109 volunteers, collected using the BCI2000 system[7]. The experimental protocol involved subjects performing various motor and imagery tasks while 64-channel EEGs were recorded. Each subject completed 14 runs:
-
Two one-minute baseline runs (eyes open and eyes closed).
-
Three two-minute runs of four different tasks:
- Task 1: Open and close left or right fist.
- Task 2: Imagine opening and closing left or right fist.
- Task 3: Open and close both fists or both feet.
- Task 4: Imagine opening and closing both fists or both feet.
The EEG signals were sampled at 160 Hz. Annotations indicate rest (T0), cue for motion or imagined motion (T1 for left fist or both fists, T2 for right fist or both feet)[6].
The data is in EDF+ format, with annotations. The EEGs were recorded from 64 electrodes following the international 10-10 system, excluding specific electrodes.[7]
The data processing involves reading EEG data from EDF files, filtering, and segmenting it into epochs. The key steps include:
-
Directory Setup: The code expects a specific directory structure with EDF files organized by subject and run.
-
Reading and Filtering: For each EDF file, the mne library is used to read the data, apply a notch filter at 60 Hz to remove powerline noise, and a high-pass filter at 2 Hz.
-
Epoch Creation:
-
For baseline data (baseline_open, baseline_closed), fixed-length epochs of 60 seconds are created.
-
For task data (task_1 to task_4), epochs are created based on annotations within the EDF file. These epochs are 4.1 seconds long, matching the duration of the tasks.
-
-
Labeling: Labels are assigned to each epoch based on the task type and event annotations.
-
Padding/Trimming: Data is padded or trimmed to ensure all epochs have the same length.
-
Saving Data: Processed data is saved as .pkl files for later use in model training.
The neural network architecture illustrated in Figure 1 is EEGNet, designed specifically for processing EEG (Electroencephalography) signals. EEGNet is widely used in brain-computer interface applications due to its efficiency and effectiveness in handling EEG data.[9] The architecture comprises three main blocks: initial convolutional layers, depthwise separable convolutional layers, and a final classification layer. [8]
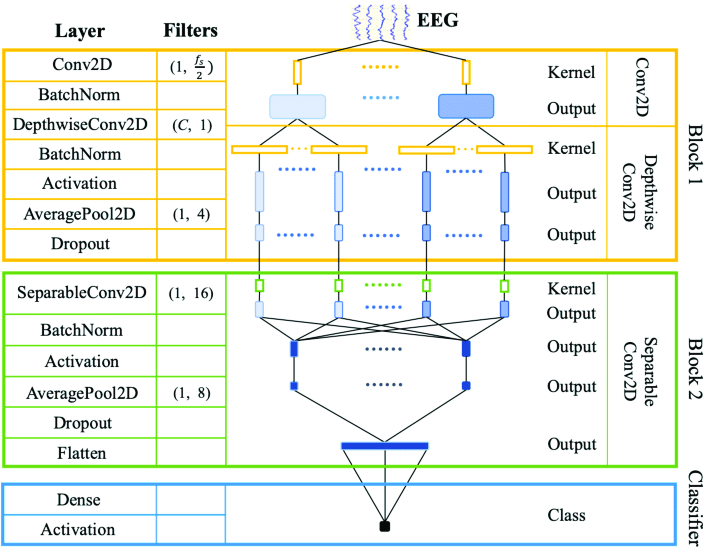 |
|---|
| Figure 1 - EEGNet |
The first block aims to capture essential spatial and temporal features from the EEG data.
-
Conv2D Layer:
- The initial layer applies a two-dimensional convolution to the input EEG data, utilizing 1 input channel and
$\frac{f_s}{2}$ output channels, where$f_s$ denotes the sampling frequency of the EEG signals.
- The initial layer applies a two-dimensional convolution to the input EEG data, utilizing 1 input channel and
-
Batch Normalization:
- Following the Conv2D layer, batch normalization is applied to stabilize and accelerate the training process by normalizing the activations.
-
DepthwiseConv2D Layer:
- This layer performs depthwise convolution with
$C$ filters, where$C$ corresponds to the number of EEG channels. This operation processes each channel independently, preserving spatial information.
- This layer performs depthwise convolution with
-
Batch Normalization:
- Another batch normalization layer is used to normalize the activations post-depthwise convolution.
-
Activation Function:
- An activation function, such as ReLU, is applied to introduce non-linearity, enabling the network to learn more complex patterns.
-
Average Pooling (AveragePool2D):
- Average pooling with a filter size of
$1 \times 4$ reduces the spatial dimensions by averaging over non-overlapping 4x4 blocks.
- Average pooling with a filter size of
-
Dropout:
- Dropout regularization is implemented to prevent overfitting by randomly setting a fraction of the input units to zero during training.
The second block is designed to further process the features extracted by the initial block in a parameter-efficient manner.
-
SeparableConv2D Layer:
- This layer employs depthwise separable convolutions with 16 filters, decomposing the convolution operation into a depthwise spatial convolution followed by a pointwise convolution, significantly reducing the computational complexity.
-
Batch Normalization:
- Batch normalization is again utilized to normalize the activations from the SeparableConv2D layer.
-
Activation Function:
- An activation function is applied to introduce additional non-linearity.
-
Average Pooling (AveragePool2D):
- Average pooling with a filter size of
$1 \times 8$ is used to further downsample the spatial dimensions.
- Average pooling with a filter size of
-
Dropout:
- Dropout regularization is included to mitigate overfitting.
-
Flatten Layer:
- This layer flattens the multi-dimensional feature maps into a one-dimensional vector, making it suitable for subsequent fully connected layers.
The final component of the architecture is the classifier, which consists of:
-
Dense Layer:
- A fully connected dense layer that consolidates the features from the previous layers to make a prediction.
-
Activation Function:
- The final activation function, typically softmax, is applied to convert the network outputs into class probabilities.
This architecture effectively balances complexity and computational efficiency, making it well-suited for processing and analyzing EEG data. By leveraging initial convolutional layers to capture spatial-temporal features, depthwise separable convolutions for efficient feature extraction, and robust regularization techniques, it achieves a high degree of performance in EEG signal classification tasks.
The model is trained using StratifiedKFold cross-validation to ensure each fold has a similar class distribution, reducing the risk of overfitting and providing a robust assessment of the model's performance. Specifically, the dataset is split into 5 folds:
The implementation of EEGNet v4 model is from braindecode.
The training process involves initializing the model weights using Glorot uniform initialization and biases set to zero. For each fold in the cross-validation, the model is trained for 400 epochs using the Adam optimizer and cross-entropy loss with a learning rate of 0.005 and batch size of 2048. After training each fold, the model's performance is evaluated on the validation set, and metrics such as accuracy, sensitivity, specificity, PPV, and NPV are calculated using confusion matrices. The trained models for each fold are saved to disk for later use and comparison. Finally, the models are evaluated on a separate test set to assess their generalization performance.
-
Initialize Model:
- Use Glorot uniform initialization for weights.
- Set biases to zero.
- EEGNet v4
- Number of channels: 64
- Number of output classes: 5
- Kernel length: 32
-
Training Configuration:
- Optimizer: Adam
- Loss Function: Cross-entropy loss
- Learning Rate: 0.005
- Batch Size: 2048
- Epochs: 400
-
Cross-Validation:
- Stratified K-Fold Cross-Validation to ensure each fold has the same proportion of each class.
- Number of Folds: 5
-
Performance Metrics:
- Accuracy
- Sensitivity
- Specificity
- PPV (Positive Predictive Value)
- NPV (Negative Predictive Value)
- Confusion Matrices
-
Save Models:
- Save the trained models for each fold to disk
-
Evaluate on Test Set:
- Assess the generalization performance of the models on a separate test set.
After training with cross-validation, the models are evaluated on a separate test set comprising subjects after S095. The same metrics (accuracy, confusion matrix, sensitivity, specificity, PPV, and NPV) are calculated to assess the model's generalization performance.
This approach ensures a comprehensive evaluation of the EEGNet v4 model across different data splits, highlighting areas for improvement and providing a reliable measure of its classification capabilities.
 |
|---|
| Figure 2 - Loss per Fold |
 |
|---|
| Figure 3 - Confusion Matrix for each fold |
 |
|---|
| Figure 4 - Predicted(Column) Normalized Confusion Matrix for each fold |
 |
|---|
| Figure 5 - Bar Charts for various metrics |
| Accuracy | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 |
|---|---|---|---|---|---|
| Rest | 70.2117 | 71.1258 | 64.43 | 72.815 | 67.0572 |
| Left fist | 71.3578 | 69.0563 | 61.9724 | 68.4753 | 69.1822 |
| Both fists | 71.176 | 69.2196 | 66.7982 | 70.8153 | 64.9463 |
| Right fist | 72.3414 | 70.1401 | 61.4416 | 72.4765 | 66.6404 |
| Both feet | 72.1773 | 70.421 | 63.1583 | 68.9371 | 66.6712 |
| Sensitivity | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 |
|---|---|---|---|---|---|
| Rest | 50.818 | 52.741 | 41.3026 | 65.5598 | 43.128 |
| Left fist | 56.2372 | 52.552 | 31.6124 | 52.8463 | 51.2638 |
| Both fists | 54.6012 | 46.9754 | 55.5203 | 56.167 | 35.94 |
| Right fist | 56.544 | 52.1739 | 32.089 | 63.5674 | 45.2607 |
| Both feet | 56.2372 | 54.8204 | 34.9484 | 50.8539 | 49.921 |
| Specificity | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 |
|---|---|---|---|---|---|
| Rest | 89.6053 | 89.5106 | 87.5574 | 80.0702 | 90.9864 |
| Left fist | 86.4783 | 85.5607 | 92.3324 | 84.1044 | 87.1005 |
| Both fists | 87.7507 | 91.4637 | 78.0762 | 85.4637 | 93.9526 |
| Right fist | 88.1389 | 88.1062 | 90.7943 | 81.3857 | 88.0202 |
| Both feet | 88.1173 | 86.0215 | 91.3682 | 87.0204 | 83.4215 |
| PPV | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 |
|---|---|---|---|---|---|
| Rest | 50.7661 | 53.861 | 48.9642 | 43.1875 | 58.209 |
| Left fist | 46.729 | 45.799 | 54.3716 | 43.4477 | 53.6364 |
| Both fists | 48.4573 | 56.0948 | 42.2612 | 47.1713 | 63.3705 |
| Right fist | 50.136 | 50.457 | 50.1863 | 44.108 | 52.3766 |
| Both feet | 49.9546 | 47.6582 | 53.9216 | 47.5177 | 46.711 |
| NPV | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 |
|---|---|---|---|---|---|
| Rest | 89.6247 | 89.0806 | 83.7689 | 90.9589 | 84.6055 |
| Left fist | 90.356 | 88.5935 | 82.3674 | 88.5299 | 85.9932 |
| Both fists | 90.1618 | 88.137 | 85.8622 | 89.4037 | 83.4388 |
| Right fist | 90.5807 | 88.8078 | 82.2245 | 90.625 | 84.6715 |
| Both feet | 90.5184 | 89.1314 | 82.9339 | 88.4555 | 85.1244 |
Fold 1
-
A significant number of "Rest" instances are misclassified as "Right fist," indicating a challenge in distinguishing these two classes.
-
Many instances of "Both fists" are misclassified into other categories, suggesting difficulty in accurately identifying this class.
-
The model struggles particularly with distinguishing "Both fists" from other classes.
Fold 2
-
The "Rest" class has a notable number of misclassifications into other classes, indicating less clarity in identifying resting state signals.
-
The "Both fists" and "Both feet" classes show significant misclassifications, which affects the model's overall accuracy and precision for these classes.
-
There is a need for improvement in accurately classifying the "Both fists" and "Both feet" classes.
Fold 3
-
The model shows a high rate of misclassification for the "Both fists" class, significantly affecting performance.
-
The "Rest" class is frequently misclassified as other classes, showing the model's difficulty in identifying resting state signals.
-
This fold demonstrates significant challenges in distinguishing between "Rest" and other movements, with "Both fists" being particularly problematic.
Fold 4
-
There is a notable confusion between the "Rest" and "Left fist" classes, which affects accuracy.
-
The "Both fists" class continues to be problematic with many misclassifications.
-
Similar issues as other folds with additional confusion between "Rest" and "Left fist" classes.
Fold 5
-
Compared to other folds, fold 5 shows lower misclassification rates, indicating better performance.
-
Despite better overall performance, there is still some confusion between the "Rest" and "Both feet" classes.
-
Fold 5 performs comparatively better but still faces challenges in distinguishing certain classes.
Common challenges observed:
-
Frequent misclassifications suggest that the model struggles to identify the resting state accurately.
-
High misclassification rates for the "Both fists" class are observed across most folds.
-
The model shows variability in performance across folds, indicating a need for more robust training and validation processes.
One potential avenue for future work is to separate the motor and imagined tasks and analyze their performance independently. This can provide insights into the specific challenges and differences in classifying motor and imagined movements. Another interesting direction for future work is to compare the results of classifying motor, imagined, and combined motor and imagined tasks. This can help identify any differences in performance and provide a better understanding of the capabilities and limitations of the model in different scenarios.
[1] Velasco, Ivan & Sipols, A. & Simon, Clara & Pastor, Luis & Bayona, Sofia. (2023). Motor imagery EEG signal classification with a multivariate time series approach. BioMedical Engineering OnLine. 22. https://doi.org/10.1186/s12938-023-01079-x.
[2] Batistić, Luka, Diego Sušanj, Domagoj Pinčić, and Sandi Ljubic. 2023. "Motor Imagery Classification Based on EEG Sensing with Visual and Vibrotactile Guidance" Sensors 23, no. 11: 5064. https://doi.org/10.3390/s23115064
[3] Alizadeh, Nazanin & Afrakhteh, Sajjad & Mosavi, M.. (2023). Deep CNN‐based classification of motor imagery tasks from EEG signals using 2D wavelet transformed images of adaptively reconstructed signals from MVMD decomposed modes. International Journal of Imaging Systems and Technology. 33. https://doi.org/10.1002/ima.22913.
[4] Lee, Y., Lee, H. J., & Tae, K. S. (2023). Classification of EEG signals related to real and imagery knee movements using deep learning for brain computer interfaces. Technology and health care : official journal of the European Society for Engineering and Medicine, 31(3), 933–942. https://doi.org/10.3233/THC-220363
[5] T. Pham, K. Adhikari and W. G. Besio, "Deep Learning-Based Classification of Finger Movements using tEEG and EEG Signals," 2023 IEEE World AI IoT Congress (AIIoT), Seattle, WA, USA, 2023, pp. 0120-0126, doi: https://doi.org/10.1109/AIIoT58121.2023.10174357.
[6] Goldberger, A., Amaral, L., Glass, L., Hausdorff, J., Ivanov, P. C., Mark, R., ... & Stanley, H. E. (2000). PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation [Online]. 101 (23), pp. e215–e220.
[7] Schalk, G., McFarland, D.J., Hinterberger, T., Birbaumer, N., Wolpaw, J.R. BCI2000: A General-Purpose Brain-Computer Interface (BCI) System. IEEE Transactions on Biomedical Engineering 51(6):1034-1043, 2004.
[8] Schalk, G., McFarland, D.J., Hinterberger, T., Birbaumer, N., Wolpaw, J.R. BCI2000: A General-Purpose Brain-Computer Interface (BCI) System. IEEE Transactions on Biomedical Engineering 51(6):1034-1043, 2004.
[9] G. Amrani, A. Adadi, M. Berrada, Z. Souirti and S. Boujraf, "EEG signal analysis using deep learning: A systematic literature review," 2021 Fifth International Conference On Intelligent Computing in Data Sciences (ICDS), Fez, Morocco, 2021, pp. 1-8, doi: 10.1109/ICDS53782.2021.9626707. keywords: {Deep learning;Systematics;Computational modeling;Bibliographies;Computer architecture;Brain modeling;Electroencephalography;Deep learning;Electroencephalography;EEG;Machine Learning;Systematic Literature Review},
# preprocess.py
import os
import numpy as np
import mne
import pickle
# Directory where the data is stored
data_dir = 'data' # Change this to your actual data directory
# Mapping of run indices to tasks/baselines
run_mapping = {
1: 'baseline_open',
2: 'baseline_closed',
3: 'task_1',
4: 'task_2',
5: 'task_3',
6: 'task_4',
7: 'task_1',
8: 'task_2',
9: 'task_3',
10: 'task_4',
11: 'task_1',
12: 'task_2',
13: 'task_3',
14: 'task_4'
}
# Mapping of event IDs to labels
event_id_mapping = {
'T0': 1, # Rest
'T1_left': 2, # Left fist
'T1_both': 3, # Both fists
'T2_right': 4, # Right fist
'T2_both': 5 # Both feet
}
def process_run(edf_path, run_index, tmin=0, tmax=4.1):
task_label = run_mapping.get(run_index, 'unknown')
raw = mne.io.read_raw_edf(edf_path, preload=True)
raw.notch_filter(freqs=60.0) # powerline notch filter
raw.filter(2, None, method='iir') # High-pass filter at 2 Hz
# Define event annotations
events, event_id = mne.events_from_annotations(raw)
labels = []
epochs_data_list = []
# Handle baselines differently since they are long T0 periods
if task_label in ['baseline_open', 'baseline_closed']:
tmin_baseline = 0 # start from the beginning
tmax_baseline = 60 # 60 seconds for baseline
epochs = mne.make_fixed_length_epochs(raw, duration=tmax_baseline, preload=True, overlap=0)
epochs_data = epochs.get_data() * 1000 # Convert to microvolts
labels = [event_id_mapping['T0']] * epochs_data.shape[0] # Label all as T0
if epochs_data.shape[0] > 0: # Only append if there are valid epochs
epochs_data_list.append(epochs_data)
else:
# Create epochs based on static tmin and tmax values
epochs = mne.Epochs(raw, events, event_id, tmin=tmin, tmax=tmax, proj=False, baseline=(0,4.1), preload=True)
epochs_data = epochs.get_data() * 1000 # Convert to microvolts
for i, event in enumerate(epochs.events):
if task_label in ['task_1', 'task_2']:
if event[2] == event_id.get('T0'):
labels.append(event_id_mapping['T0'])
elif event[2] == event_id.get('T1'):
labels.append(event_id_mapping['T1_left'])
elif event[2] == event_id.get('T2'):
labels.append(event_id_mapping['T2_right'])
elif task_label in ['task_3', 'task_4']:
if event[2] == event_id.get('T0'):
labels.append(event_id_mapping['T0'])
elif event[2] == event_id.get('T1'):
labels.append(event_id_mapping['T1_both'])
elif event[2] == event_id.get('T2'):
labels.append(event_id_mapping['T2_both'])
else:
labels.append(event_id_mapping['T0'])
print(labels)
if epochs_data.shape[0] > 0: # Only append if there are valid epochs
epochs_data_list.append(epochs_data)
return epochs_data_list, labels, task_label
def pad_or_trim(data, target_length):
def pad(array):
if array.shape[2] < target_length:
pad_width = target_length - array.shape[2]
return np.pad(array, ((0, 0), (0, 0), (0, pad_width)), mode='constant')
return array[:, :, :target_length]
return np.concatenate([pad(d) for d in data], axis=0)
def save_data(data_dir, tmin=0, tmax=4.1):
subjects = [d for d in os.listdir(data_dir) if os.path.isdir(os.path.join(data_dir, d))]
max_samples = int((tmax - tmin) * 160) # Assuming a sampling rate of 160 Hz
baseline_samples = 60 * 160 # Assuming 60 seconds for baselines
for subject in subjects:
subject_dir = os.path.join(data_dir, subject)
edf_files = [f for f in os.listdir(subject_dir) if f.endswith('.edf')]
for edf_file in edf_files:
edf_path = os.path.join(subject_dir, edf_file)
run_index = int(edf_file.split('R')[1].split('.')[0]) # Extract run index from filename
epochs_data_list, labels, task_label = process_run(edf_path, run_index, tmin, tmax)
if len(labels) > 0 and epochs_data_list: # Only process if there are labels and epochs
if task_label in ['baseline_open', 'baseline_closed']:
padded_data = pad_or_trim(epochs_data_list, baseline_samples)
else:
padded_data = pad_or_trim(epochs_data_list, max_samples)
labels = np.array(labels)
# Create the save directory
save_dir = os.path.join('preprocessed_data', subject, task_label)
os.makedirs(save_dir, exist_ok=True)
# Save the data for each run
save_path = os.path.join(save_dir, f"run_{run_index}.pkl")
with open(save_path, 'wb') as f:
pickle.dump({'X': padded_data, 'y': labels}, f)
print(f"Saved {subject} - {task_label} - run {run_index} data.")
# Save preprocessed data for all subjects
save_data(data_dir)
print("All preprocessed data saved.")# -*- coding: utf-8 -*-
"""iter_3.ipynb
Automatically generated by Colab.
Original file is located at
https://colab.research.google.com/drive/1AGGrjq0ox8C4qWjQE8mAKLnu1PLTkr7u
"""
from google.colab import drive
drive.mount('/content/drive')
!nvidia-smi
import multiprocessing
num_cores = multiprocessing.cpu_count()
print(f"Number of CPU cores: {num_cores}")
!pip3 install torch --upgrade -q
!pip install numpy scikit-learn matplotlib visualkeras braindecode==0.8.1 skorch torchviz -q
# !python /content/drive/MyDrive/bme442/bme442project/preprocess.py
import numpy as np
import pickle
import mne
import pandas as pd
import torch
import torch.optim as optim
from torch import nn
from torch.nn.functional import elu
from torch.utils.data import DataLoader, Dataset
from torchsummary import summary
from torchviz import make_dot
from braindecode.models.base import EEGModuleMixin, deprecated_args
from braindecode.models.functions import squeeze_final_output
from braindecode.models.modules import Ensure4d, Expression
from skorch import NeuralNetClassifier
from skorch.callbacks import LRScheduler, TrainEndCheckpoint
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay, accuracy_score
from einops.layers.torch import Rearrange
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import matplotlib as mpl
import os
import concurrent.futures
import logging
import time
from tqdm import tqdm
import seaborn as sns
cuda = torch.cuda.is_available()
logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s')
base_dir = '/content/drive/MyDrive/bme442/bme442project'
preprocessed_data_dir = os.path.join(base_dir, 'preprocessed_data')
# someList = []
# with open('/content/drive/MyDrive/bme442/bme442project/preprocessed_data/S001/task_2/run_4.pkl', 'rb') as f:
# data = pickle.load(f)
# print('X: ', data['X'])
# # someList.append(data['X'][28])
# print('y: ', data['y'])
# # print(someList)
def process_run(run_path, test_flag):
logging.info(f"Processing file: {run_path}")
with open(run_path, 'rb') as f:
data = pickle.load(f)
# Get indices of all `1`s in `data['y']`
one_indices = [i for i, y in enumerate(data['y']) if y == 1]
# Create a set of indices to filter out: 4 out of every 5 `1`s
filter_indices = set(one_indices[1::5] + one_indices[2::5] + one_indices[3::5] + one_indices[4::5])
# Filter data
filtered_X = [x for i, x in enumerate(data['X']) if i not in filter_indices]
filtered_y = [y for i, y in enumerate(data['y']) if i not in filter_indices]
if test_flag:
return filtered_X, filtered_y, 'test'
else:
return filtered_X, filtered_y, 'train'
def process_subject(subject):
global test_flag
subject_dir = os.path.join(preprocessed_data_dir, subject)
tasks = ['task_1', 'task_2', 'task_3', 'task_4']
results = []
if subject == 'S095':
test_flag = True
if os.path.isdir(subject_dir):
for task in tasks:
task_dir = os.path.join(subject_dir, task)
if os.path.isdir(task_dir):
for run_file in os.listdir(task_dir):
if run_file.endswith('.pkl'):
run_path = os.path.join(task_dir, run_file)
results.append((run_path, test_flag))
return results
test_flag = False
X_combined = []
y_combined = []
X_combined_test = []
y_combined_test = []
all_runs = []
counter = 0
# Process all subjects concurrently
with concurrent.futures.ThreadPoolExecutor() as subject_executor:
subject_futures = {subject_executor.submit(process_subject, subject): subject for subject in os.listdir(preprocessed_data_dir)}
for future in tqdm(concurrent.futures.as_completed(subject_futures), total=len(subject_futures), desc="Processing subjects"):
try:
subject_runs = future.result()
all_runs.extend(subject_runs)
except Exception as e:
subject = subject_futures[future]
print(f"Error processing subject {subject}: {e}")
# Process all runs concurrently
with concurrent.futures.ThreadPoolExecutor() as executor:
futures = []
for run_path, flag in all_runs:
futures.append(executor.submit(process_run, run_path, flag))
for future in tqdm(concurrent.futures.as_completed(futures), total=len(futures), desc="Processing runs"):
filtered_X, filtered_y, dataset_type = future.result()
if dataset_type == 'test':
X_combined_test.extend(filtered_X)
y_combined_test.extend(filtered_y)
else:
X_combined.extend(filtered_X)
y_combined.extend(filtered_y)
# Subtract one from every y value
y_combined = [y - 1 for y in y_combined]
y_combined_test = [y - 1 for y in y_combined_test]
y_combined = np.array(y_combined)
X_combined = np.array(X_combined)
y_combined_test = np.array(y_combined_test)
X_combined_test = np.array(X_combined_test)
y_combined.shape, X_combined.shape, y_combined_test.shape, X_combined_test.shape
print("X_combined shape:", X_combined.shape)
print("y_combined shape:", y_combined.shape)
unique, counts = np.unique(y_combined, return_counts=True)
print(dict(zip(unique, counts)))
if X_combined.shape[0] != y_combined.shape[0]:
raise ValueError("Mismatch between number of samples in X_combined and y_combined.")
if X_combined_test.shape[0] != y_combined_test.shape[0]:
raise ValueError("Mismatch between number of samples in X_combined_test and y_combined_test.")
class Conv2dWithConstraint(nn.Conv2d):
def __init__(self, *args, max_norm=1, **kwargs):
self.max_norm = max_norm
super(Conv2dWithConstraint, self).__init__(*args, **kwargs)
def forward(self, x):
self.weight.data = torch.renorm(
self.weight.data, p=2, dim=0, maxnorm=self.max_norm
)
return super(Conv2dWithConstraint, self).forward(x)
def _glorot_weight_zero_bias(model):
"""Initialize parameters of all modules by initializing weights with
glorot
uniform/xavier initialization, and setting biases to zero. Weights from
batch norm layers are set to 1.
Parameters
----------
model: Module
"""
for module in model.modules():
if hasattr(module, "weight"):
if "BatchNorm" not in module.__class__.__name__:
nn.init.xavier_uniform_(module.weight, gain=1)
else:
nn.init.constant_(module.weight, 1)
if hasattr(module, "bias"):
if module.bias is not None:
nn.init.constant_(module.bias, 0)
# Set up logging configuration
logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s')
logger = logging.getLogger(__name__)
class EEGNetv4(EEGModuleMixin, nn.Sequential):
"""EEGNet v4 model from Lawhern et al 2018.
See details in [EEGNet4]_.
Parameters
----------
final_conv_length : int | "auto"
If int, final length of convolutional filters.
in_chans :
Alias for n_chans.
n_classes:
Alias for n_outputs.
input_window_samples :
Alias for n_times.
Notes
-----
This implementation is not guaranteed to be correct, has not been checked
by original authors, only reimplemented from the paper description.
References
----------
.. [EEGNet4] Lawhern, V. J., Solon, A. J., Waytowich, N. R., Gordon,
S. M., Hung, C. P., & Lance, B. J. (2018).
EEGNet: A Compact Convolutional Network for EEG-based
Brain-Computer Interfaces.
arXiv preprint arXiv:1611.08024.
"""
def __init__(
self,
n_chans=None,
n_outputs=None,
n_times=None,
final_conv_length="auto",
pool_mode="mean",
F1=8,
D=2,
F2=16, # usually set to F1*D (?)
kernel_length=64,
third_kernel_size=(8, 4),
drop_prob=0.25,
chs_info=None,
input_window_seconds=None,
sfreq=None,
in_chans=None,
n_classes=None,
input_window_samples=None,
):
n_chans, n_outputs, n_times = deprecated_args(
self,
("in_chans", "n_chans", in_chans, n_chans),
("n_classes", "n_outputs", n_classes, n_outputs),
("input_window_samples", "n_times", input_window_samples, n_times),
)
super().__init__(
n_outputs=n_outputs,
n_chans=n_chans,
chs_info=chs_info,
n_times=n_times,
input_window_seconds=input_window_seconds,
sfreq=sfreq,
)
del n_outputs, n_chans, chs_info, n_times, input_window_seconds, sfreq
del in_chans, n_classes, input_window_samples
if final_conv_length == "auto":
assert self.n_times is not None
self.final_conv_length = final_conv_length
self.pool_mode = pool_mode
self.F1 = F1
self.D = D
self.F2 = F2
self.kernel_length = kernel_length
self.third_kernel_size = third_kernel_size
self.drop_prob = drop_prob
# For the load_state_dict
# When padronize all layers,
# add the old's parameters here
self.mapping = {
"conv_classifier.weight": "final_layer.conv_classifier.weight",
"conv_classifier.bias": "final_layer.conv_classifier.bias",
}
pool_class = dict(max=nn.MaxPool2d, mean=nn.AvgPool2d)[self.pool_mode]
self.add_module("ensuredims", Ensure4d())
self.add_module("dimshuffle", Rearrange("batch ch t 1 -> batch 1 ch t"))
self.add_module(
"conv_temporal",
nn.Conv2d(
1,
self.F1,
(1, self.kernel_length),
stride=1,
bias=False,
padding=(0, self.kernel_length // 2),
),
)
self.add_module(
"bnorm_temporal",
nn.BatchNorm2d(self.F1, momentum=0.01, affine=True, eps=1e-3),
)
self.add_module(
"conv_spatial",
Conv2dWithConstraint(
self.F1,
self.F1 * self.D,
(self.n_chans, 1),
max_norm=1,
stride=1,
bias=False,
groups=self.F1,
padding=(0, 0),
),
)
self.add_module(
"bnorm_1",
nn.BatchNorm2d(self.F1 * self.D, momentum=0.01, affine=True, eps=1e-3),
)
self.add_module("elu_1", Expression(elu))
self.add_module("pool_1", pool_class(kernel_size=(1, 4), stride=(1, 4)))
self.add_module("drop_1", nn.Dropout(p=self.drop_prob))
# https://discuss.pytorch.org/t/how-to-modify-a-conv2d-to-depthwise-separable-convolution/15843/7
self.add_module(
"conv_separable_depth",
nn.Conv2d(
self.F1 * self.D,
self.F1 * self.D,
(1, 16),
stride=1,
bias=False,
groups=self.F1 * self.D,
padding=(0, 16 // 2),
),
)
self.add_module(
"conv_separable_point",
nn.Conv2d(
self.F1 * self.D,
self.F2,
(1, 1),
stride=1,
bias=False,
padding=(0, 0),
),
)
self.add_module(
"bnorm_2",
nn.BatchNorm2d(self.F2, momentum=0.01, affine=True, eps=1e-3),
)
self.add_module("elu_2", Expression(elu))
self.add_module("pool_2", pool_class(kernel_size=(1, 8), stride=(1, 8)))
self.add_module("drop_2", nn.Dropout(p=self.drop_prob))
output_shape = self.get_output_shape()
n_out_virtual_chans = output_shape[2]
if self.final_conv_length == "auto":
n_out_time = output_shape[3]
self.final_conv_length = n_out_time
# Incorporating classification module and subsequent ones in one final layer
module = nn.Sequential()
module.add_module(
"conv_classifier",
nn.Conv2d(
self.F2,
self.n_outputs,
(n_out_virtual_chans, self.final_conv_length),
bias=True,
),
)
if self.add_log_softmax:
module.add_module("logsoftmax", nn.LogSoftmax(dim=1))
# Transpose back to the logic of braindecode,
# so time in third dimension (axis=2)
module.add_module(
"permute_back",
Rearrange("batch x y z -> batch x z y"),
)
module.add_module("squeeze", Expression(squeeze_final_output))
self.add_module("final_layer", module)
_glorot_weight_zero_bias(self)
def forward(self, x):
logger.info(f'Input shape: {x.shape}')
x = super().forward(x)
logger.info(f'Output shape: {x.shape}')
return x
# Ensure device is correctly set
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# Step 1: Check for NaN or infinite values in the data
assert not np.isnan(X_combined).any(), "X_combined contains NaN values"
assert not np.isinf(X_combined).any(), "X_combined contains infinite values"
assert not np.isnan(y_combined).any(), "y_combined contains NaN values"
assert not np.isinf(y_combined).any(), "y_combined contains infinite values"
# Step 2: Convert to PyTorch tensors on CPU first
X_combined_tensor_cpu = torch.from_numpy(X_combined).float()
y_combined_tensor_cpu = torch.from_numpy(y_combined).long()
# Check the tensors on CPU
print(f"Shape of X_combined_tensor_cpu: {X_combined_tensor_cpu.shape}")
print(f"Shape of y_combined_tensor_cpu: {y_combined_tensor_cpu.shape}")
# Test with a small batch
small_batch_X = X_combined_tensor_cpu[:10]
small_batch_y = y_combined_tensor_cpu[:10]
try:
small_batch_X_cuda = small_batch_X.to(device)
small_batch_y_cuda = small_batch_y.to(device)
print("Small batch transfer successful")
except RuntimeError as e:
print(f"Error during small batch transfer: {e}")
# Inspect the data for unusual values
print("X_combined max:", X_combined.max())
print("X_combined min:", X_combined.min())
print("X_combined mean:", X_combined.mean())
print("y_combined unique values:", np.unique(y_combined))
# Enable CUDA debugging mode
os.environ['CUDA_LAUNCH_BLOCKING'] = "1"
# Move the tensors to the CUDA device again
try:
X_combined_tensor = X_combined_tensor_cpu.to(device)
y_combined_tensor = y_combined_tensor_cpu.to(device)
print("Data transfer to CUDA successful")
except RuntimeError as e:
print(f"Error during data transfer to CUDA: {e}")
def process_fold(train_index, test_index, X_combined, y_combined, batch_size=128):
print("process_fold called")
assert torch.cuda.is_available(), "CUDA is not available. Check your GPU setup."
torch.backends.cudnn.enabled = False # Disable cuDNN for debugging purposes
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
logger.info(f"Using device: {device}")
X_train, X_test = X_combined[train_index], X_combined[test_index]
y_train, y_test = y_combined[train_index], y_combined[test_index]
# Check for NaN or infinite values in the data
assert not np.isnan(X_train).any(), "X_train contains NaN values"
assert not np.isinf(X_train).any(), "X_train contains infinite values"
assert not np.isnan(X_test).any(), "X_test contains NaN values"
assert not np.isinf(X_test).any(), "X_test contains infinite values"
assert not np.isnan(y_train).any(), "y_train contains NaN values"
assert not np.isinf(y_train).any(), "y_train contains infinite values"
assert not np.isnan(y_test).any(), "y_test contains NaN values"
assert not np.isinf(y_test).any(), "y_test contains infinite values"
# Convert data to PyTorch tensors
X_train_tensor_cpu = torch.from_numpy(X_train).float()
X_test_tensor_cpu = torch.from_numpy(X_test).float()
y_train_tensor_cpu = torch.from_numpy(y_train).long()
y_test_tensor_cpu = torch.from_numpy(y_test).long()
# Transfer full tensors to the CUDA device
X_train = X_train_tensor_cpu.to(device)
X_test = X_test_tensor_cpu.to(device)
y_train = y_train_tensor_cpu.to(device)
y_test = y_test_tensor_cpu.to(device)
# Define the model
model = EEGNetv4(n_chans=64, n_outputs=5, n_times=656, kernel_length=32)
model = model.to(device)
# Define the loss function and optimizer
criterion = nn.CrossEntropyLoss()
optimizer = optim.Adam(model.parameters(), lr=0.005, capturable=False) # capturable=False to avoid CUDA assertion
# Training loop
def train_model(model, criterion, optimizer, X_train, y_train, n_epochs=400, batch_size=128):
model.train()
dataset = torch.utils.data.TensorDataset(X_train, y_train)
dataloader = torch.utils.data.DataLoader(dataset, batch_size=batch_size, shuffle=True)
loss_history = []
for epoch in range(n_epochs):
epoch_start_time = time.time()
running_loss = 0.0
for batch_idx, (inputs, labels) in enumerate(dataloader):
batch_start_time = time.time()
inputs, labels = inputs.to(device), labels.to(device)
optimizer.zero_grad()
outputs = model(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
batch_end_time = time.time()
print(f'Batch {batch_idx + 1}/{len(dataloader)} processed in {batch_end_time - batch_start_time:.4f} seconds')
avg_loss = running_loss / len(dataloader)
loss_history.append(avg_loss)
epoch_end_time = time.time()
logger.info(f'Epoch [{epoch + 1}/{n_epochs}] completed in {epoch_end_time - epoch_start_time:.4f} seconds, Average Loss: {avg_loss:.4f}')
return model, loss_history
logger.info('Starting model training...')
print('Starting model training...')
model, loss_history = train_model(model, criterion, optimizer, X_train, y_train, batch_size=batch_size)
logger.info('Model training completed.')
print('Model training completed.')
# Predict and evaluate
logger.info('Starting prediction...')
print('Starting prediction...')
model.eval()
with torch.no_grad():
outputs = model(X_test)
_, y_pred = torch.max(outputs, 1)
y_pred = y_pred.cpu().numpy()
accuracy = accuracy_score(y_test.cpu().numpy(), y_pred)
logger.info(f'Fold accuracy: {accuracy:.4f}')
print(f'Fold accuracy: {accuracy:.4f}')
# Generate confusion matrix
cm = confusion_matrix(y_test.cpu().numpy(), y_pred)
logger.info(f'Confusion Matrix:\n{cm}')
print(f'Confusion Matrix:\n{cm}')
# Calculate sensitivities, specificities, PPVs, and NPVs
fold_sensitivities = []
fold_specificities = []
fold_ppvs = []
fold_npvs = []
for i in range(5):
tp = cm[i, i]
fn = cm[i, :].sum() - tp
fp = cm[:, i].sum() - tp
tn = cm.sum() - (tp + fn + fp)
sensitivity = tp / (tp + fn) if (tp + fn) > 0 else 0
specificity = tn / (tn + fp) if (tn + fp) > 0 else 0
ppv = tp / (tp + fp) if (tp + fp) > 0 else 0
npv = tn / (tn + fn) if (tn + fn) > 0 else 0
fold_sensitivities.append(sensitivity)
fold_specificities.append(specificity)
fold_ppvs.append(ppv)
fold_npvs.append(npv)
logger.info(f'Fold sensitivities: {fold_sensitivities}')
print(f'Fold sensitivities: {fold_sensitivities}')
logger.info(f'Fold specificities: {fold_specificities}')
print(f'Fold specificities: {fold_specificities}')
logger.info(f'Fold PPVs: {fold_ppvs}')
print(f'Fold PPVs: {fold_ppvs}')
logger.info(f'Fold NPVs: {fold_npvs}')
print(f'Fold NPVs: {fold_npvs}')
return model, fold_sensitivities, fold_specificities, fold_ppvs, fold_npvs, cm, loss_history
skf = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
all_sensitivities = []
all_specificities = []
all_ppvs = []
all_npvs = []
models = []
cms = []
loss_histories = []
# Check if the trained_models directory exists and create if it doesn't
os.makedirs(os.path.join(base_dir, 'trained_models'), exist_ok=True)
# Check for existing models
existing_models = os.listdir(os.path.join(base_dir, "trained_models"))
if existing_models:
print("Loading existing models...")
for fold_index, model_file in enumerate(existing_models, start=1):
model_path = os.path.join(base_dir, 'trained_models', model_file)
model = EEGNetv4(n_chans=64, n_outputs=5, n_times=656, kernel_length=32)
model.load_state_dict(torch.load(model_path))
models.append(model)
print(f"Loaded model for fold {fold_index}")
else:
print("Starting StratifiedKFold cross-validation")
fold_index = 1
for train_index, test_index in skf.split(X_combined, y_combined):
print(f"Processing fold {fold_index}")
model, sensitivities, specificities, ppvs, npvs, cm, loss_history = process_fold(train_index, test_index, X_combined, y_combined, batch_size=2048)
if sensitivities and specificities and ppvs and npvs:
all_sensitivities.append(sensitivities)
all_specificities.append(specificities)
all_ppvs.append(ppvs)
all_npvs.append(npvs)
models.append(model)
cms.append(cm)
loss_histories.append(loss_history)
model_save_path = os.path.join(base_dir, 'trained_models', f'model_fold_{fold_index}.pth')
torch.save(model.state_dict(), model_save_path)
print(f"Completed fold {fold_index}")
fold_index += 1
print("Completed StratifiedKFold cross-validation")
# Convert lists to numpy arrays for further analysis
all_sensitivities = np.array(all_sensitivities)
all_specificities = np.array(all_specificities)
all_ppvs = np.array(all_ppvs)
all_npvs = np.array(all_npvs)
print(f'Mean Sensitivities per fold: {all_sensitivities.mean(axis=0)}')
print(f'Mean Specificities per fold: {all_specificities.mean(axis=0)}')
print(f'Mean PPVs per fold: {all_ppvs.mean(axis=0)}')
print(f'Mean NPVs per fold: {all_npvs.mean(axis=0)}')
# Plot loss graphs fold by fold
def plot_loss_graphs(loss_histories):
fig, ax = plt.subplots()
for fold, loss_history in enumerate(loss_histories, start=1):
ax.plot(loss_history, label=f'Fold {fold}')
ax.set_xlabel('Epochs')
ax.set_ylabel('Loss')
ax.set_title('Training Loss per Fold')
ax.legend()
plt.show()
plot_loss_graphs(loss_histories)
# Test the resulting models with y_combined_test and X_combined_test
X_combined_test_tensor = torch.from_numpy(X_combined_test).float().to(device)
y_combined_test_tensor = torch.from_numpy(y_combined_test).long().to(device)
all_test_sensitivities = []
all_test_specificities = []
all_test_ppvs = []
all_test_npvs = []
test_cms = []
cms = []
for i, model in enumerate(models, 1):
model.to(device) # Ensure the model is on the same device as the input
model.eval()
with torch.no_grad():
outputs = model(X_combined_test_tensor)
_, y_pred = torch.max(outputs, 1)
y_pred = y_pred.cpu().numpy()
cm = confusion_matrix(y_combined_test, y_pred)
cms.append(cm)
test_cms.append(cm)
print(f'Confusion Matrix for model {i}:\n{cm}')
# torch.save(model.state_dict(), os.path.join(base_dir, 'trained_models', f'model_test_{i}.pth'))
# Calculate sensitivities, specificities, PPVs, and NPVs
test_sensitivities = []
test_specificities = []
test_ppvs = []
test_npvs = []
for j in range(len(cm)): # Adjusted to use len(cm) for the range
tp = cm[j, j]
fn = cm[j, :].sum() - tp
fp = cm[:, j].sum() - tp
tn = cm.sum() - (tp + fn + fp)
sensitivity = tp / (tp + fn) if (tp + fn) > 0 else 0
specificity = tn / (tn + fp) if (tn + fp) > 0 else 0
ppv = tp / (tp + fp) if (tp + fp) > 0 else 0
npv = tn / (tn + fn) if (tn + fn) > 0 else 0
test_sensitivities.append(sensitivity)
test_specificities.append(specificity)
test_ppvs.append(ppv)
test_npvs.append(npv)
all_test_sensitivities.append(test_sensitivities)
all_test_specificities.append(test_specificities)
all_test_ppvs.append(test_ppvs)
all_test_npvs.append(test_npvs)
print("Completed testing models on the test set")
# Convert the confusion matrices to pandas DataFrames
df_cms = []
for i, cm in enumerate(cms, 1):
df_cm = pd.DataFrame(cm, columns=[f'Predicted Class {i}' for i in range(5)], index=[f'Actual Class {i}' for i in range(5)])
df_cms.append(df_cm)
# Define the class names
class_names = ['Rest', 'Left fist', 'Both fists', 'Right fist', 'Both feet']
# Create a figure with subplots for each fold, adjusting layout and size
fig, axes = plt.subplots(1, len(df_cms), figsize=(20, 8), constrained_layout=True)
# Plot each confusion matrix
for i, df_cm in enumerate(df_cms, 1):
sns.heatmap(df_cm, annot=True, fmt='g', cmap='Blues', ax=axes[i-1], xticklabels=class_names, yticklabels=class_names)
axes[i-1].set_xlabel('Predicted Class')
axes[i-1].set_ylabel('Actual Class')
axes[i-1].set_title(f'Fold {i} Confusion Matrix')
axes[i-1].tick_params(axis='x', rotation=45)
axes[i-1].tick_params(axis='y', rotation=45)
plt.show()
all_test_sensitivities = np.array(all_test_sensitivities)
all_test_specificities = np.array(all_test_specificities)
all_test_ppvs = np.array(all_test_ppvs)
all_test_npvs = np.array(all_test_npvs)
all_test_accuracies = (all_test_sensitivities + all_test_specificities) / 2
print(f'Mean Test Sensitivities: {all_test_sensitivities.mean(axis=0)}')
print(f'Mean Test Specificities: {all_test_specificities.mean(axis=0)}')
print(f'Mean Test PPVs: {all_test_ppvs.mean(axis=0)}')
print(f'Mean Test NPVs: {all_test_npvs.mean(axis=0)}')
# Convert to DataFrames with class names as indices
df_sensitivities = pd.DataFrame(all_test_sensitivities * 100, index=class_names, columns=[f'Fold {i+1}' for i in range(all_test_sensitivities.shape[1])])
df_specificities = pd.DataFrame(all_test_specificities * 100, index=class_names, columns=[f'Fold {i+1}' for i in range(all_test_specificities.shape[1])])
df_ppvs = pd.DataFrame(all_test_ppvs * 100, index=class_names, columns=[f'Fold {i+1}' for i in range(all_test_ppvs.shape[1])])
df_npvs = pd.DataFrame(all_test_npvs * 100, index=class_names, columns=[f'Fold {i+1}' for i in range(all_test_npvs.shape[1])])
df_accuracies = pd.DataFrame(all_test_accuracies * 100, index=class_names, columns=[f'Fold {i+1}' for i in range(all_test_accuracies.shape[1])])
# Print the tables
print("Test Sensitivities by Fold")
print(df_sensitivities)
print("\nTest Specificities by Fold")
print(df_specificities)
print("\nTest PPVs by Fold")
print(df_ppvs)
print("\nTest NPVs by Fold")
print(df_npvs)
print("\nTest Accuracies by Fold")
print(df_accuracies)
# List of DataFrames and their corresponding titles
dataframes = [
(df_accuracies, 'Test Accuracies by Fold'),
(df_sensitivities, 'Test Sensitivities by Fold'),
(df_specificities, 'Test Specificities by Fold'),
(df_ppvs, 'Test PPVs by Fold'),
(df_npvs, 'Test NPVs by Fold')
]
# Define the function to display bar charts in a custom grid
def display_combined_bar_charts(dataframes):
fig = plt.figure(figsize=(20, 12))
spec = gridspec.GridSpec(ncols=6, nrows=2, figure=fig)
axes = []
axes.append(fig.add_subplot(spec[0, 0:2])) # row 0, spans columns 0-1
axes.append(fig.add_subplot(spec[0, 2:4])) # row 0, spans columns 2-3
axes.append(fig.add_subplot(spec[0, 4:])) # row 0, spans columns 4-5
axes.append(fig.add_subplot(spec[1, 1:3])) # row 1, spans columns 1-2
axes.append(fig.add_subplot(spec[1, 3:5])) # row 1, spans columns 3-4
for ax, (df, title) in zip(axes, dataframes):
df.plot(kind='bar', ax=ax)
ax.set_title(title)
ax.set_xlabel('Fold')
ax.set_ylabel('Percentage')
ax.legend(title='Metrics', bbox_to_anchor=(1.05, 1), loc='upper left')
ax.set_xticklabels(df.index, rotation=0)
plt.tight_layout()
plt.show()
display_combined_bar_charts(dataframes)
df_cms = [np.array(cm) for cm in df_cms]
# Normalize the confusion matrices along the columns
df_cms_normalized = [cm.astype('float') / cm.sum(axis=0)[np.newaxis, :] for cm in df_cms]
# Define the class names
class_names = ['Rest', 'Left fist', 'Both fists', 'Right fist', 'Both feet']
# Create a figure with subplots for each fold, adjusting layout and size
fig, axes = plt.subplots(1, len(df_cms_normalized), figsize=(20, 8), constrained_layout=True)
# Plot each normalized confusion matrix
for i, df_cm in enumerate(df_cms_normalized, 1):
sns.heatmap(df_cm, annot=True, fmt='.2f', cmap='Blues', ax=axes[i-1], xticklabels=class_names, yticklabels=class_names)
axes[i-1].set_xlabel('Predicted Class')
axes[i-1].set_ylabel('Actual Class')
axes[i-1].set_title(f'Fold {i} Confusion Matrix')
axes[i-1].tick_params(axis='x', rotation=45)
axes[i-1].tick_params(axis='y', rotation=45)
plt.show()