Welcome to the APAeval GitHub repository.
APAeval is a community effort to evaluate computational methods for the detection and quantification of poly(A) sites and the estimation of their differential usage across RNA-seq samples.
APAeval consists of three challenges, each consisting of a set of benchmarks for tools that:
- Identify polyadenylation sites
- Quantify polyadenylation sites
- Calculate differential usage of polyadenylation sites
For more info, please refer to our landing page.
If you would like to contribute to APAeval, the first things you would need to do are:
- Register for one or more of the APAeval teams
- Please use this form to provide us with your user names/handles of the various service accounts we are using (see below) so that we can add you to the corresponding repositories/organizations
- Wait for us to reach out to you with an invitation to our Slack space, then
use the invitation to sign up and post a short intro message in the
#generalchannel
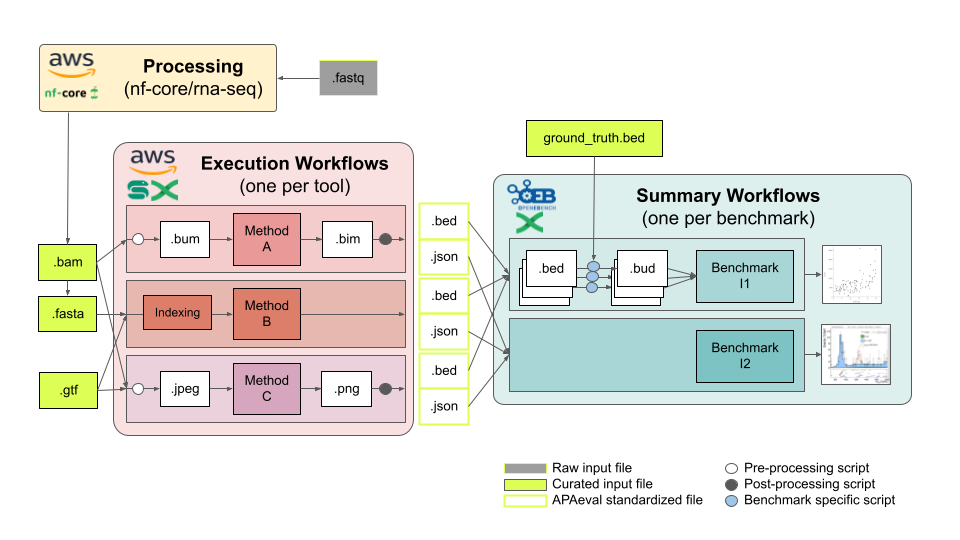
The bulk of the work falls into roughly two tasks, writing method execution workflows and benchmark summary workflows as highlighted in the figure below:
Execution workflows contain all steps that need to be run per method:
- Pre-processing: Convert the input files the APAeval team has prepared into the input files a given method consumes, if applicable.
- Method execution: Execute the method in any way necessary to compute the output files for all challenges (may require more than one run of the tool if, e.g., run in different execution modes).
- Post-processing: Convert the output files of the method into the formats consumed by the summary workflows as specified by the APAeval team, if applicable.
Execution workflows should be implemented in either Nexflow or Snakemake, and individual steps should be isolated through the use of either Conda virtual environments (deprecated; to run on AWS we need containerized workflows) or Docker/Singularity containers.
Summary workflows contain all steps that need to be run per benchmark, using outputs of the invididual method execution workflows as inputs. They follow the OpenEBench workflow model, described here, implemented in Nextflow. OpenEBench workflows consist of the following 3 steps:
- Validation: Output data of the various execution workflows is validated against the provided specifications to data consistency.
- Metrics computation: Actual benchmarking metrics are computed as specified, e.g., by comparisons to ground truth/gold standard data sets.
- Consolidation: Data is consolidated for consistency with other benchmarking efforts, based on the OpenEBench/ELIXIR benchmarking data model.
Following the OpenEBench workflows model also ensures that result visualizations are automatically generated, as long as the desired graph types are supported by OpenEBench.
Apart from writing execution and summary workflows, there are various other smaller jobs that participants can work on, including, e.g.:
- Pre-processing RNA-Seq input data via the nf-core RNA-Seq analysis pipeline
- Writing additional benchmark specifications
- Housekeeping jobs (reviewing code, helping to keep the repository clean, enforce good coding practices, etc.)
- Work with our partner OpenEBench on their open issues, e.g., by extending their portfolio of supported visualization
If you do not know where to start, simply ask us!
To account for everyone's different agendas and time zones, the hackathon is organized such that contributors can work, as much as possible, in their own time.
Following best practices for writing software and sharing data and code is important to us, and therefore we want to apply, as much as possible, FAIR Principles to data and software alike. This includes publishing all code open source, under permissive licenses approved by the Open Source Initiative and all data by a permissive Creative Commons license.
In particular, we publish all code under the MIT license and all data under the CC0 license. An exception are all summary workflows, which are published under the GPLv3 license, as the provided template is derived from an OpenEBench example workflow that is itself licensed under GPLv3. A copy of the MIT license is also shipped with this repository.
We also believe that attribution, provenance and transparency are crucial for an open and fair work environment in the sciences, especially in a community effort like APAeval. Therefore, we would like to make clear from the beginning that in all publications deriving from APAeval (journal manuscript, data and code repositories), any non-trivial contributions will be acknowledged by authorship. All authors will be strictly listed alphabetically, by last name, with no exceptions, wherever possible under the name of The APAeval Team and accompanied by a more detailed description of how people contributed.
We expect that all contributors accept the license and attribution policies outlined above.
We are making use of Slack (see above to see how you can become a member) for asynchronous communication. Please use the most appropriate channels for discussions/questions etc.:
#general: Introduce yourself and find pointers to get you started. All APAeval-wide announcements will be put here!#admin: Ask questions about the general organization of APAeval.#tech-support: Ask questions about the technical infrastructure and relevant software, e.g., AWS, GitHub, Nextflow, Snakemake.#challenge-differential-usage: Discuss the poly(A) site differential usage challenge.#challenge-identification: Discuss the poly(A) site identification challenge.#challenge-quantification: Discuss the poly(A) site quantification challenge.#tool-{METHOD_NAME}: Discussions channels for each indidvidual method.#random: Post anything that doesn't fit into any of the other channels.
Despite the event taking place mostly asynchrounously, we do have a few video calls to increase the feeling of collaboration. In particular, we hae a kickoff and a wrap-up meeting, three reporting calls and the presentation of APAeval during the RNA Meeting. Please join meetings if you can, we have tried to shuffle the times a bit, so that most people should be able to attend at least some of the calls, irrespective of your location.
This calendar contains all video call events, including the necessary login info, and we would like to kindly ask you to subscribed to it:
- Calendar ID:
[email protected] - Public address
Please do not download the ICS file and then import it, as any updates to the calendar will not be synced. Instead, copy the calendar ID or public address and paste it in the appropriate field of your calendar application. Refer to your calendar application's help pages if you do not know how to subscribeh to a calendar.
With the exception of presenting APAeval during the RNA Meeting, all video calls will take place in the following Zoom room:
- Direct link
- Meeting ID:
656 9429 1427 - Passcode:
APAeval
There is also a meeting agenda.
For more lively meetings, participants are encouraged to switch on their cameras. But please mute your microphones if you are not currently speaking.
We are making extensive use of GitHub's project management resources to allow contributors to work independently on individual, largely self-contained, issues. There are several Kanban project boards, listing relevant issues for different kinds of tasks, such as drafting benchmarking specifications and implementing/running method execution workflows.
The idea is that people assign themselves to open issues (i.e., those issues that are not yet assigned to someone else). Note that in order to be able to do so, you will need to be a member of this GitHub repository (see above to see how you can become a member). Once you have assigned yourself, you can move/drag the issue from the To do to the In progress column of the Kanban board.
When working on an issue, please start by cloning (preferred) or forking the
repository. Then create a feature branch and implement your code/changes. Once
you are happy with them, please create a pull request against the main
branch, making sure to fill in the provided template (in particular, please
refer to the original issue you are addressing with this pull request) and to
assign two reviewers. This workflow ensures collaborative coding and is
sometimes referred to as GitHub flow. If you are not familiar with
Git, GitHub or the GitHub flow, there are many useful tutorials online, e.g.,
those listed below.
AWS kindly sponsored credits for their compute and storage infrastructure that we can use to run any heavy duty computations in the cloud (e.g., RNA-Seq data pre-processing by or method execution workflows).
This also includes credits to run Seqara Lab's Nextflow Tower, a convenient web-based platform to run Nextflow workflows, such as the nf-core RNA-Seq analysis workflow we are using for pre-processing RNA-Seq data. Seqera Labs has kindly offered to hold a workshop on Nextflow and Nextflow Tower and will be providing technical support during the hackathon.
Setting up the AWS organization and infrastructure is still ongoing, and we will update this section with more information as soon as that is done.
We are partnering with OpenEBench, a benchmarking and technical monitoring platform for bioinformatics tools. OpenEBench development, maintenance and operation is coordinated by Barcelona Supercomputing Center (BSC) together with partners from the European Life Science infrastructure initiative ELIXIR.
OpenEBench tooling will facilitate the computation and visualization of benchmarking results and store the results of all challenges and benchmarks in their databases, making it easy for others to explore results. This should also make it easy to add additional methods to existing benchmarking challenges later on. OpenEBench developers are also advising us on creating benchmarks that are compatible with good practices in the wider community of bioinformatics challenges.
The OpenEBench team will give a presentation to APAeval participants on how the service is structured and what it offers to its visitors and the APAeval effort.
Here are some pointers and tutorials for the main software tools that we are going to use throughout the hackathon:
- Conda: tutorial
- Docker: tutorial
- Git: tutorial
- GitHub: general tutorial / GitHub flow tutorial
- Nextflow: tutorial
- Singularity: tutorial
- Snakemake: tutorial
Note that you don't need to know about all of these, e.g., one of Conda (deprecated; to run on AWS we need containerized workflows), Docker and/or Singularity will typically be enough. See below, for a discussion of the supported workflow languages/management systems. Again, working with one will be enough for most issues.
In addition to these, basic programming/scripting skills may be required for most, but not for all issues. For those that do, you are generally free to choose your preferred language, although for those people who have experience with Python, we recommend you to go with that. It just makes it easier for others to review your code, and it typically integrates better with our templates and the general bioinformatics ecosystem/community.
Note that even if you don't have experience with any of these tools/languages, and you don't feel like or have no time learning them, there is surely still something that you can help us with. Just ask us and we will try to put your individual skills to good use! 💪
As mentioned further above, we would like execution workflows to be written in one of two "workflow languages": Nextflow or Snakemake. Specifying workflows in such a language rather than, say, by stringing together Bash commands, is considered good practice, as it increases reusability and reproducibility, and so is in line with our goal of adhering to FAIR software principles.
But why only Nextflow and Snakemake, and not, e.g., the Common Workflow Language, the Workflow Definition Language or Galaxy? There are no particular reasons other than that APAeval organizers have experience with these workflows languages and are thus able to provide technical support. If you are an experienced workflow developer and prefer another workflow language, you are welcome to use that one instead, but note that we have no templates available and will not be able to help you much in case you encounter issues during development or execution.
As for summary workflows, we are bound to implement these in Nextflow, as they are executed on OpenEBench, which currently only accepts Nextflow workflows.
For this reason, as well as the fact that we will provide Nextflow Tower for convient execution of Nextflow workflows on AWS cloud infrastructure (see above) and use a Nextflow analysis pipeline for pre-processing RNA-Seq data sets, we recommend novices without any other considerations (e.g., colleagues already working with Snakemake) to use Nextflow.
Please be kind to one another and mind the Contributor Covenant's Code of Conduct for all interactions with the community. A copy of the Code of Conduct is also shipped with this repository. Please report any violations to the Code of Conduct to either or both of Christina and Alex via Slack.
Thanks goes to these wonderful people (emoji key):
Chelsea Herdman 📆 |
This project follows the all-contributors specification. Contributions of any kind welcome!