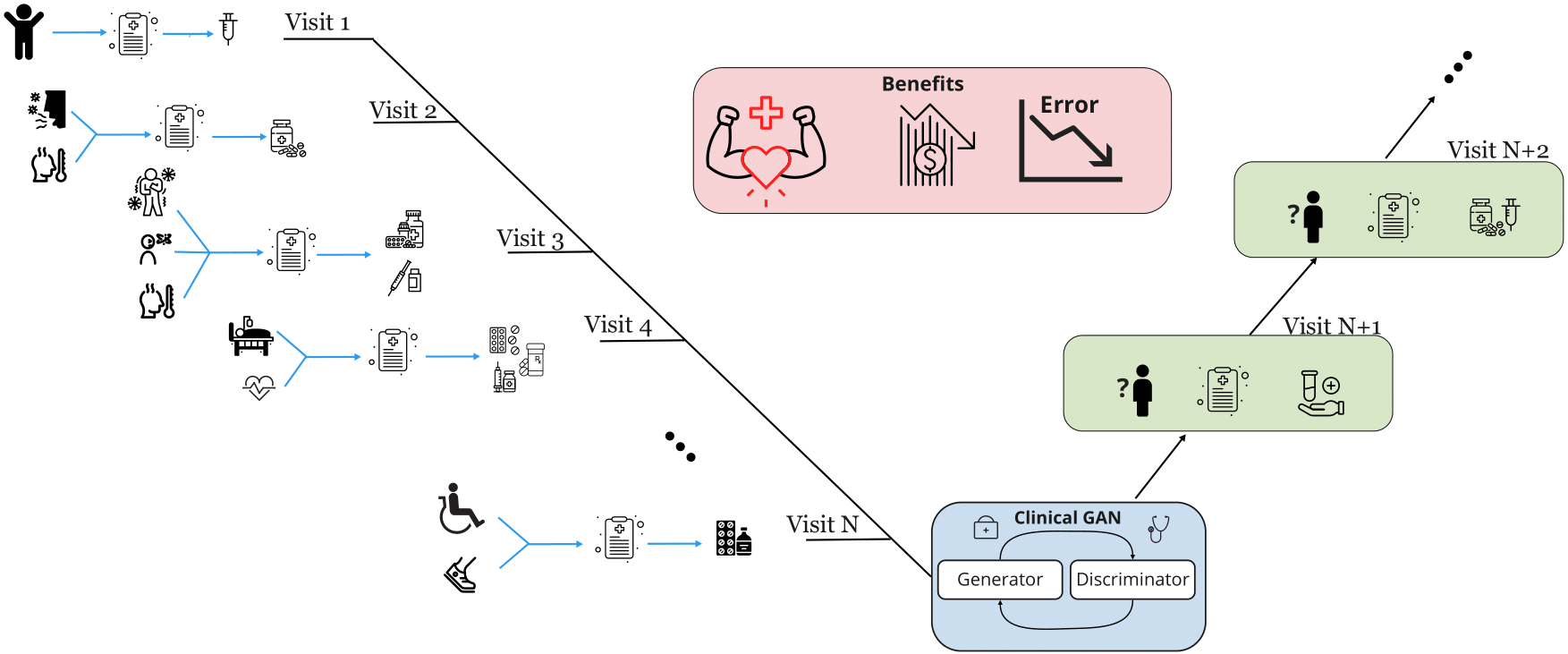
Clinical-GAN: Trajectory forecasting model for clinical events using Transformer and Generative Adversarial Network (paper)
Predicting the trajectory of a disease at an early stage can aid physicians in offering effective treatment, prompt care to patients, and also avoid misdiagnosis. However, forecasting patient trajectories is challenging due to long-range dependencies, irregular intervals between consecutive admissions, and non-stationarity data. To address these challenges, we propose a novel method called Clinical-GAN, a Transformer-based Generative Adversarial Networks (GAN) to forecast the patients’ medical codes for subsequent visits. First, we represent the patients’ medical codes as a time-ordered sequence of tokens akin to language models. Then, a Transformer mechanism is used as a Generator to learn from existing patients’ medical history and is trained adversarially against a Transformer-based Discriminator. We address the above mentioned challenges based on our data modeling and Transformer-based GAN architecture. Additionally, we enable the local interpretation of the model’s prediction using a multi-head attention mechanism. We evaluated our method using a publicly available dataset, Medical Information Mart for Intensive Care IV v1.0 (MIMIC-IV), with more than 500,000 visits completed by around 196,000 adult patients over an 11-year period from 2008–2019. Clinical-GAN significantly outperforms baseline methods and existing works, as demonstrated through various experiments.
The Requirement.txt file lists all Python libraries that this project needs, install using the below command.
pip install -r Requirement.txt
Please install postgresql 13.1 from postgresql.
We do not provide the MIMIC-IV data. In order to access the data, one must be an credentialied user on Physionet. Please visit physionet for more instructions. We also do not provide the CCS and CCSR mapping data. Please visit HCUP to download CCS (Single level CCS) , CCSR for Diagnosis and CCSR for procedure files. After that, Move the downloaded folders to the current working directory.
/Clinical-GAN/DXCCSR_v2021-2
/Clinical-GAN/PRCCSR_v2021-1
/Clinical-GAN/Single_Level_CCS_2015
Replace the path in QUERIES.SQL to the current working directory. More specifically, replace the following current_working_directory to your path where you have stored this project.
Please do not change the filenames.
COPY 'query' To current_working_directory/Clinical-GAN/data/*.csv
To prepare the data as mentioned in the paper, Run the following command:
python process_data.py
Following are the optional arguments.
--mimic3_path: Path of mimic IV CSV files where the queried data is stored. Default is 'data'.--CCSRDX_file: Path of diagnosis based CCSR files. Default is 'DXCCSR_v2021-2/DXCCSR_v2021-2.csv'--CCSRPCS_file: Path of procedure based CCSR files. Default is 'PRCCSR_v2021-1/PRCCSR_v2021-1.csv'--D_CCSR_Ref_file: Path of diagnosis based CCSR Reference file. Default is 'DXCCSR_v2021-2/DXCCSR-Reference-File-v2021-2.xlsx'--P_CCSR_Ref_file: Path of procedure based CCSR Reference file. Default is 'PRCCSR_v2021-1/PRCCSR-Reference-File-v2021-1.xlsx'--CCSDX_file: Path of diagnosis based CCS files. Default is 'Single_Level_CCS_2015/$dxref 2015.csv'--CCSPX_file: Path of procedure based CCS files. Default is 'Single_Level_CCS_2015/$prref 2015.csv'--min_dx: Minimum diagnosis code assigned per visit. Default is 80--min_px: Minimum procedure code assigned per visit. Default is 80--min_drg: Minimum drug/medication code assigned per visit. Default is 80--threshold: Remove the code whose frequency is less than the threshold. Default is 5--seqLength: maximum sequence length of each sequence in input and output. Default is 500
To train the model, Run the following command:
python train.py --task TF --scenario S1 --fileName myAwesomeModel.pt
Following are the required arguments.
--scenario: which type of scenario based data needs to be loaded- S1 (FD), S2 (FDP), S3 (FDPR). Scenarios as mentioned in the paper. Default=S1--task: Two types of task SDP and TF. Default=TF--fileName: fileName for the model which is going to be stored in the 'model' folder. Default=myAwesomeModel.pt
Following are the optional arguments involving hyperparameters.
--learning_rate: learning rate of the model. Default=4e-4.--epochs: Total number of epochs. Default=100--gen_layers: Total number of generator's Encoder and Decoder layers. Default=3--disc_layers: Total number of discriminator's Encoder layers. Default=1--dropout: Dropout value to be applied forreducing overfitting. Default=0.1--clip: Discriminator's cliping value for gradient clipping. Default=0.1--gen_clip:Generator's cliping value for gradient clipping. Default=1.0--alpha:alpha value for Generator's loss. Default=0.3--gen_heads: Total number of multi-head in Generator. Default=8--disc_heads:Total number of multi-head in Discriminator. Default=4.--batch_size: batch size to be used for training the model. Default=8--isdataparallel: if you have more than one gpu, you could use dataparallization. Default=False--hid_dim: Embedding dimension of both Generator and discriminator. Default=256--pf_dim: Hidden dimension of both Generator and discriminator. Default=512--warmup_steps: warmp up steps for learning rate. Default=30--labelSmoothing:label smoothing value for reducing overfitting. Default=0.0--factor: factor by which the learning rate value should increase or decrease. Default=1--checkpoint_dir: If you want to run the model for more epochs after terminating the training, Provide the path of the saved model. Default=None--valid_data_ratio:How much data should be allocated to valid set in percentage. Default=0.05--test_data_ratio: How much data should be allocated to test set in percentage. Default=0.05
To evaluate the model, Run the following command:
eval.py --task TF --scenario S1 --fileName myAwesomeModel.pt
Following are the required arguments
--scenario: Which type of scenario based data needs to be loaded- S1 (FD), S2 (FDP), S3 (FDPR). Scenarios as mentioned in the paper. Default=S1--task: Two types of task SDP and TF. Default=TF--fileName:Load the saved model from the 'models/ClinicalGAN' folder. Default=myAwesomeModel.pt
For Inference, Run the following command:
infer.py --task TF --scenario S1 --modelFileName myAwesomeModel
Following are the required arguments
--scenario: Which type of scenario based data needs to be loaded- S1 (FD), S2 (FDP), S3 (FDPR). Scenarios as mentioned in the paper. Default=S1--task: Two types of task SDP and TF. Default=TF--fileName:Load the saved model from the 'models/ClinicalGAN' folder. Default=myAwesomeModel.pt
Few samples forecasted by Clinical-GAN can be found here
Also check out the paper, and please cite it if you use in your work.
@article{Shankar_ClinicalGAN,
title = {Clinical-GAN: Trajectory Forecasting of Clinical Events using Transformer and Generative Adversarial Networks},
journal = {Artificial Intelligence in Medicine},
volume = {138},
pages = {102507},
year = {2023},
issn = {0933-3657},
doi = {https://doi.org/10.1016/j.artmed.2023.102507},
url = {https://www.sciencedirect.com/science/article/pii/S0933365723000210},
author = {Vignesh Shankar and Elnaz Yousefi and Alireza Manashty and Dayne Blair and Deepika Teegapuram},
}