This project depends on a customized george, setup can be done via conda:
conda create -n add python=3.11
conda activate add
conda install -y -c conda-forge libstdcxx-ng
conda install -y -c anaconda pyyaml jinja2 numpy scipy pybind11
git clone https://github.com/maxc01/addtree
cd addtree
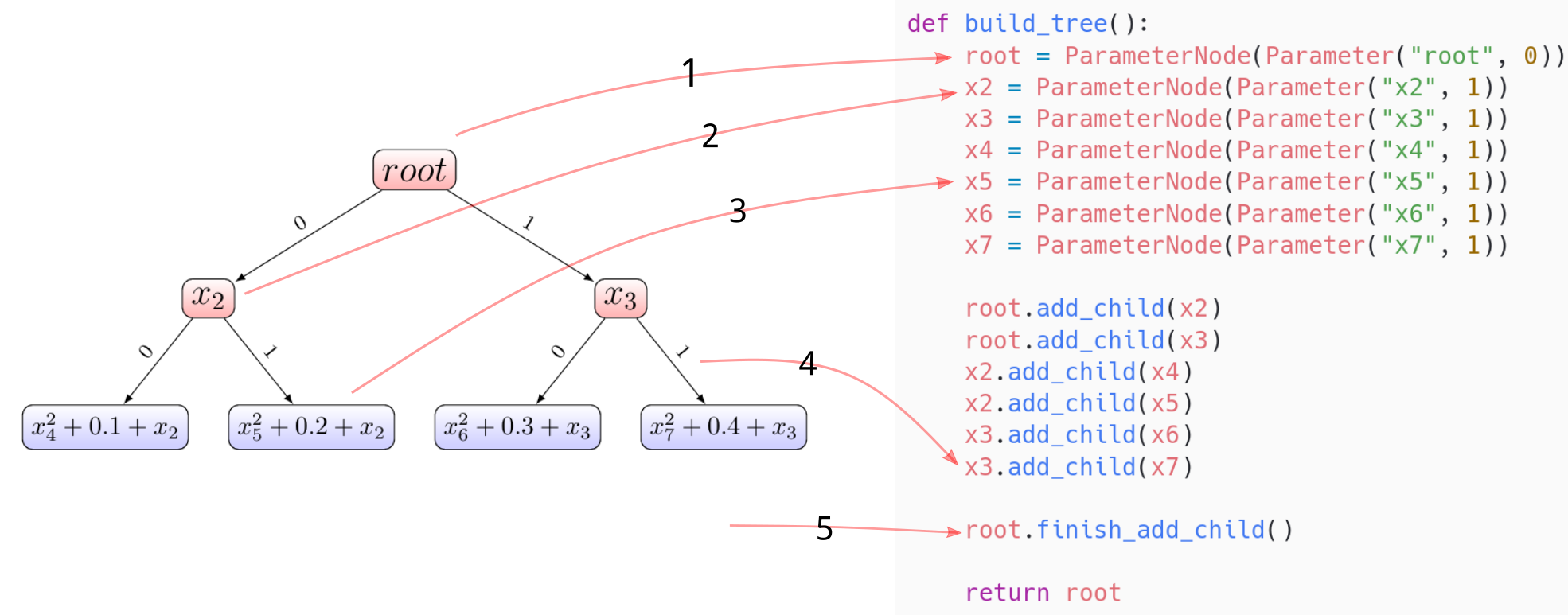
pip install .The first step is to represent an objective function at hand using a tree. For example, the synthetic function presented in Jenatton2017 can be encoded using a tree shown in the left part of the following figure and its corresponding code snippet is shown in the right part. We clarify this code snippet in the following.
rootnode doesn’t contain a continuous parameter, thus the dimension of this parameter is set to be zerox2node contains a one-dimensional continuous parameter, and this parameter is shared by two sub-functions associated with the first and second leaves (counting from left to right)x5node contains a one-dimensional continuous parameter- build tree dependencies by adding
x7as a child ofx3 - finish tree construction (precompute some quantities)
The general rule of coding a tree-structured function is using a hierarchical dictionary as the parameter of this function. For example, the code snippet corresponding to the above tree-structured function is shown below.
def obj_func(params):
SHIFT = 0.5
if params["L1"]["cat_value"] == "left" and params["L2"]["cat_value"] == "left":
value = (
params["L1"]["cont_value"] + (params["L2"]["cont_value"] - SHIFT) ** 2 + 0.1
)
elif params["L1"]["cat_value"] == "left" and params["L2"]["cat_value"] == "right":
value = (
params["L1"]["cont_value"] + (params["L2"]["cont_value"] - SHIFT) ** 2 + 0.2
)
elif params["L1"]["cat_value"] == "right" and params["L2"]["cat_value"] == "left":
value = (
params["L1"]["cont_value"] + (params["L2"]["cont_value"] - SHIFT) ** 2 + 0.3
)
elif params["L1"]["cat_value"] == "right" and params["L2"]["cat_value"] == "right":
value = (
params["L1"]["cont_value"] + (params["L2"]["cont_value"] - SHIFT) ** 2 + 0.4
)
else:
raise ValueError("parameter names are not correct")
info = dict()
info["value"] = value
info["value_sigma"] = 1e-9
return infoFor a more complex tree-structured function, see the objective function of a simple NAS problem.
cd examples
python addtree_jenatton_small.pylog files will be located in
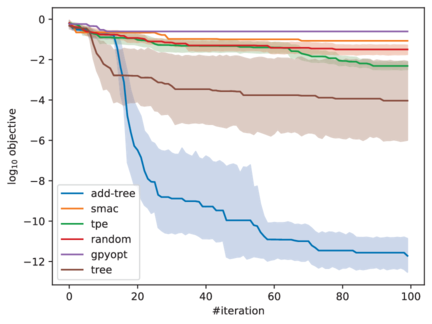
exp_results/addtree/jenatton-small/a-unique-dir. The following figure shows
comparison of different algorithms on optimizaing the synthetic function.
cd examples
python {{ algo }}_model_compression_multiple.py {{ model_name }} OUTPUT_PATH --pretrained PRETRAINED_PATH --prune_epochs 1model_name can be “vgg16”, “resnet50” or “resnet56”.
algo can be “addtree”, “random” , “tpe” or “smac”.
For example, to compress resnet50 using addtree,
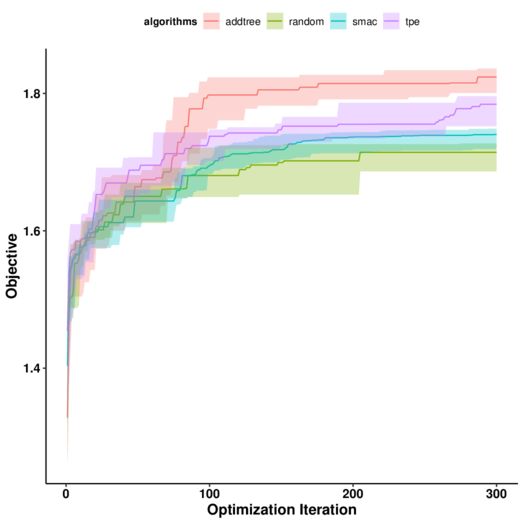
python addtree_model_compression_multiple.py resnet50 OUTPUT_PATH --pretrained PRETRAINED_PATH --prune_epochs 1The following picture shows comparison of different algorithms on compressing resnet50.
@inproceedings{ma2020,
title = {Additive {{Tree}}-{{Structured Covariance Function}} for {{Conditional Parameter Spaces}} in {{Bayesian Optimization}}},
booktitle = {International {{Conference}} on {{Artificial Intelligence}} and {{Statistics}}},
author = {Ma, Xingchen and Blaschko, Matthew},
year = {2020},
month = jun,
pages = {1015--1025},
publisher = {{PMLR}},
issn = {2640-3498},
language = {en}
}
@article{ma2020b,
title = {Additive Tree-Structured Conditional Parameter Spaces in Bayesian Optimization: {{A}} Novel Covariance Function and a Fast Implementation},
author = {Ma, X. and Blaschko, M. B.},
year = {2020},
publisher = {{IEEE Computer Society}},
address = {{Los Alamitos, CA, USA}},
issn = {1939-3539},
doi = {10.1109/TPAMI.2020.3026019},
journal = {IEEE Transactions on Pattern Analysis \& Machine Intelligence},
keywords = {additives,bayes methods,data models,linear programming,mathematical model,neural networks,optimization}
}