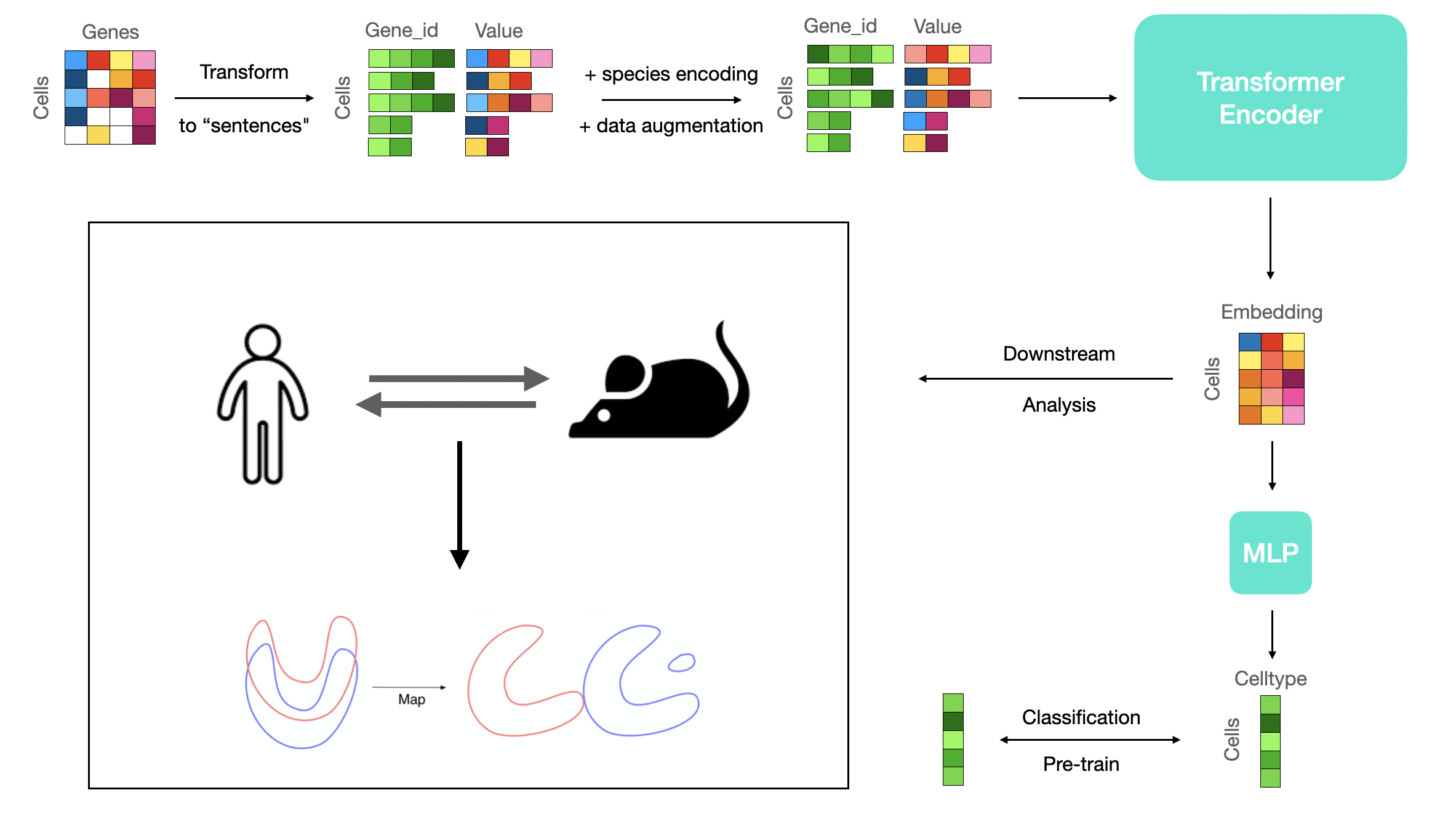
scTT: Translating Transcriptional Human-Mouse Single-Cell Signatures using Transformer-based Representational Learning
import torch
from module import ScTT
from dataset import SingleDataset
from torch.utils.data import DataLoader
sctt = ScTT(
n_genes = 10000,
n_val = 11,
n_class = 1000,
n_species=2
pooling='mean',
embed_dim=768,
n_heads=8,
n_layers=8,
lr=1e-4,
)
dataset = SingleDataset(adata,gene2id)
loader = DataLoader(dataset)
sctt.fit(model, loader)n_genes: int.
Amount of genes.n_val: int.
Max expression value after preprocessing.n_class: int.
Number of classes to classify.n_species: int.
Number of species.embed_dim: int.
Dimension of the embeddings.n_heads: int, default8.
Number of heads in Multi-head Attention layer.n_layers: int, default8.
Number of Transformer blocks.lr: float between[0, 1], default0.0001.
Learning rate.pooling: string, eithermaxpooling,minpooling ormeanpooling
Please consider citing the following reference:
https://icml-compbio.github.io/icml-website-2020/2020/papers/WCBICML2020_paper_29.pdf https://www.biorxiv.org/content/10.1101/2020.02.05.935239v2