Documentation (Latest | Stable) | DGL at a glance | Model Tutorials | Official Examples | Discussion Forum | Slack Channel
For a full list of official DGL examples, see here.
DGL is an easy-to-use, high performance and scalable Python package for deep learning on graphs. DGL is framework agnostic, meaning if a deep graph model is a component of an end-to-end application, the rest of the logics can be implemented in any major frameworks, such as PyTorch, Apache MXNet or TensorFlow.

Figure: DGL Overall Architecture
02/26/2021: The new v0.6.0 release includes distributed heterogeneous graph support, 13 more model examples, a Chinese translation of user guide thank to community support, and a new tutorial. See our release note for more details.
09/05/2020: We invite you to participate in the survey here to make DGL better fit for your needs. Thanks!
08/21/2020: The new v0.5.0 release includes distributed GNN training, overhauled documentation and user guide, and several more features. We have also submitted some models to the OGB leaderboard. See our release note for more details.
A data scientist may want to apply a pre-trained model to your data right away. For this you can use DGL's Application packages, formally Model Zoo. Application packages are developed for domain applications, as is the case for DGL-LifeScience. We will soon add model zoo for knowledge graph embedding learning and recommender systems. Here's how you will use a pretrained model:
from dgllife.data import Tox21
from dgllife.model import load_pretrained
from dgllife.utils import smiles_to_bigraph, CanonicalAtomFeaturizer
dataset = Tox21(smiles_to_bigraph, CanonicalAtomFeaturizer())
model = load_pretrained('GCN_Tox21') # Pretrained model loaded
model.eval()
smiles, g, label, mask = dataset[0]
feats = g.ndata.pop('h')
label_pred = model(g, feats)
print(smiles) # CCOc1ccc2nc(S(N)(=O)=O)sc2c1
print(label_pred[:, mask != 0]) # Mask non-existing labels
# tensor([[ 1.4190, -0.1820, 1.2974, 1.4416, 0.6914,
# 2.0957, 0.5919, 0.7715, 1.7273, 0.2070]])Further reading: DGL is released as a managed service on AWS SageMaker, see the medium posts for an easy trip to DGL on SageMaker(part1 and part2).
Researchers can start from the growing list of models implemented in DGL. Developing new models does not mean that you have to start from scratch. Instead, you can reuse many pre-built modules. Here is how to get a standard two-layer graph convolutional model with a pre-built GraphConv module:
from dgl.nn.pytorch import GraphConv
import torch.nn.functional as F
# build a two-layer GCN with ReLU as the activation in between
class GCN(nn.Module):
def __init__(self, in_feats, h_feats, num_classes):
super(GCN, self).__init__()
self.gcn_layer1 = GraphConv(in_feats, h_feats)
self.gcn_layer2 = GraphConv(h_feats, num_classes)
def forward(self, graph, inputs):
h = self.gcn_layer1(graph, inputs)
h = F.relu(h)
h = self.gcn_layer2(graph, h)
return hNext level down, you may want to innovate your own module. DGL offers a succinct message-passing interface (see tutorial here). Here is how Graph Attention Network (GAT) is implemented (complete codes). Of course, you can also find GAT as a module GATConv:
import torch.nn as nn
import torch.nn.functional as F
# Define a GAT layer
class GATLayer(nn.Module):
def __init__(self, in_feats, out_feats):
super(GATLayer, self).__init__()
self.linear_func = nn.Linear(in_feats, out_feats, bias=False)
self.attention_func = nn.Linear(2 * out_feats, 1, bias=False)
def edge_attention(self, edges):
concat_z = torch.cat([edges.src['z'], edges.dst['z']], dim=1)
src_e = self.attention_func(concat_z)
src_e = F.leaky_relu(src_e)
return {'e': src_e}
def message_func(self, edges):
return {'z': edges.src['z'], 'e':edges.data['e']}
def reduce_func(self, nodes):
a = F.softmax(nodes.mailbox['e'], dim=1)
h = torch.sum(a * nodes.mailbox['z'], dim=1)
return {'h': h}
def forward(self, graph, h):
z = self.linear_func(h)
graph.ndata['z'] = z
graph.apply_edges(self.edge_attention)
graph.update_all(self.message_func, self.reduce_func)
return graph.ndata.pop('h')Microbenchmark on speed and memory usage: While leaving tensor and autograd functions to backend frameworks (e.g. PyTorch, MXNet, and TensorFlow), DGL aggressively optimizes storage and computation with its own kernels. Here's a comparison to another popular package -- PyTorch Geometric (PyG). The short story is that raw speed is similar, but DGL has much better memory management.
| Dataset | Model | Accuracy | Time PyG DGL |
Memory PyG DGL |
|---|---|---|---|---|
| Cora | GCN GAT |
81.31 ± 0.88 83.98 ± 0.52 |
0.478 0.666 1.608 1.399 |
1.1 1.1 1.2 1.1 |
| CiteSeer | GCN GAT |
70.98 ± 0.68 69.96 ± 0.53 |
0.490 0.674 1.606 1.399 |
1.1 1.1 1.3 1.1 |
| PubMed | GCN GAT |
79.00 ± 0.41 77.65 ± 0.32 |
0.491 0.690 1.946 1.393 |
1.1 1.1 1.6 1.1 |
| GCN | 93.46 ± 0.06 | OOM 28.6 | OOM 11.7 | |
| Reddit-S | GCN | N/A | 29.12 9.44 | 15.7 3.6 |
Table: Training time(in seconds) for 200 epochs and memory consumption(GB)
Here is another comparison of DGL on TensorFlow backend with other TF-based GNN tools (training time in seconds for one epoch):
| Dateset | Model | DGL | GraphNet | tf_geometric |
|---|---|---|---|---|
| Core | GCN | 0.0148 | 0.0152 | 0.0192 |
| GCN | 0.1095 | OOM | OOM | |
| PubMed | GCN | 0.0156 | 0.0553 | 0.0185 |
| PPI | GCN | 0.09 | 0.16 | 0.21 |
| Cora | GAT | 0.0442 | n/a | 0.058 |
| PPI | GAT | 0.398 | n/a | 0.752 |
High memory utilization allows DGL to push the limit of single-GPU performance, as seen in below images.
 |
 |
|---|
Scalability: DGL has fully leveraged multiple GPUs in both one machine and clusters for increasing training speed, and has better performance than alternatives, as seen in below images.
 |
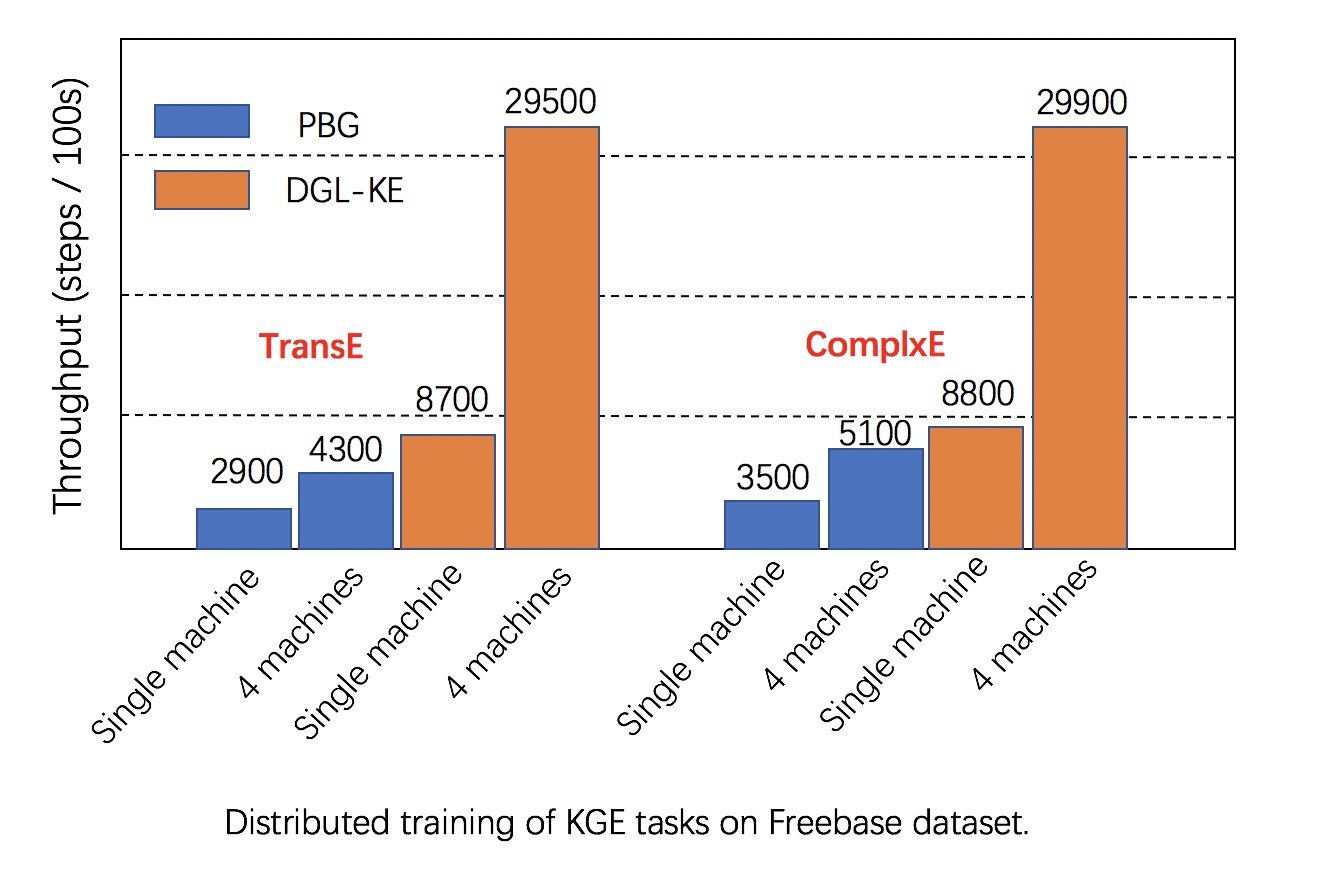 |
|---|
Further reading: Detailed comparison of DGL and other Graph alternatives can be found here.
Overall there are 30+ models implemented by using DGL:
- DGL-LifeSci, previously DGL-Chem
- DGL-KE
- DGL-RecSys(coming soon)
We are currently in Beta stage. More features and improvements are coming.
-
Benchmarking Graph Neural Networks, Vijay Prakash Dwivedi, Chaitanya K. Joshi, Thomas Laurent, Yoshua Bengio, Xavier Bresson
-
Open Graph Benchmarks: Datasets for Machine Learning on Graphs, NeurIPS'20, Weihua Hu, Matthias Fey, Marinka Zitnik, Yuxiao Dong, Hongyu Ren, Bowen Liu, Michele Catasta, Jure Leskovec
-
DropEdge: Towards Deep Graph Convolutional Networks on Node Classification, ICLR'20, Yu Rong, Wenbing Huang, Tingyang Xu, Junzhou Huan
-
Discourse-Aware Neural Extractive Text Summarization, ACL'20, Jiacheng Xu, Zhe Gan, Yu Cheng, Jingjing Liu
-
GCC: Graph Contrastive Coding for Graph Neural Network Pre-Training, KDD'20, Jiezhong Qiu, Qibin Chen, Yuxiao Dong, Jing Zhang, Hongxia Yang, Ming Ding, Kuansan Wang, Jie Tang
-
DGL-KE: Training Knowledge Graph Embeddings at Scale, SIGIR'20, Da Zheng, Xiang Song, Chao Ma, Zeyuan Tan, Zihao Ye, Jin Dong, Hao Xiong, Zheng Zhang, George Karypis
-
Improving Graph Neural Network Expressivity via Subgraph Isomorphism Counting, Giorgos Bouritsas, Fabrizio Frasca, Stefanos Zafeiriou, Michael M. Bronstein
-
INT: An Inequality Benchmark for Evaluating Generalization in Theorem Proving, Yuhuai Wu, Albert Q. Jiang, Jimmy Ba, Roger Grosse
-
Finding Patient Zero: Learning Contagion Source with Graph Neural Networks, Chintan Shah, Nima Dehmamy, Nicola Perra, Matteo Chinazzi, Albert-László Barabási, Alessandro Vespignani, Rose Yu
-
FeatGraph: A Flexible and Efficient Backend for Graph Neural Network Systems, SC'20, Yuwei Hu, Zihao Ye, Minjie Wang, Jiali Yu, Da Zheng, Mu Li, Zheng Zhang, Zhiru Zhang, Yida Wang
more
-
BP-Transformer: Modelling Long-Range Context via Binary Partitioning., Zihao Ye, Qipeng Guo, Quan Gan, Xipeng Qiu, Zheng Zhang
-
OptiMol: Optimization of Binding Affinities in Chemical Space for Drug Discovery, Jacques Boitreaud,Vincent Mallet, Carlos Oliver, Jérôme Waldispühl
-
JAKET: Joint Pre-training of Knowledge Graph and Language Understanding, Donghan Yu, Chenguang Zhu, Yiming Yang, Michael Zeng
-
Architectural Implications of Graph Neural Networks, Zhihui Zhang, Jingwen Leng, Lingxiao Ma, Youshan Miao, Chao Li, Minyi Guo
-
Combining Reinforcement Learning and Constraint Programming for Combinatorial Optimization, Quentin Cappart, Thierry Moisan, Louis-Martin Rousseau1, Isabeau Prémont-Schwarz, and Andre Cire
-
Therapeutics Data Commons: Machine Learning Datasets and Tasks for Therapeutics (code repo), Kexin Huang, Tianfan Fu, Wenhao Gao, Yue Zhao, Yusuf Roohani, Jure Leskovec, Connor W. Coley, Cao Xiao, Jimeng Sun, Marinka Zitnik
-
Sparse Graph Attention Networks, Yang Ye, Shihao Ji
-
On Self-Distilling Graph Neural Network, Yuzhao Chen, Yatao Bian, Xi Xiao, Yu Rong, Tingyang Xu, Junzhou Huang
-
Learning Robust Node Representations on Graphs, Xu Chen, Ya Zhang, Ivor Tsang, and Yuangang Pan
-
Recurrent Event Network: Autoregressive Structure Inference over Temporal Knowledge Graphs, Woojeong Jin, Meng Qu, Xisen Jin, Xiang Ren
-
Graph Neural Ordinary Differential Equations, Michael Poli, Stefano Massaroli, Junyoung Park, Atsushi Yamashita, Hajime Asama, Jinkyoo Park
-
FusedMM: A Unified SDDMM-SpMM Kernel for Graph Embedding and Graph Neural Networks, Md. Khaledur Rahman, Majedul Haque Sujon, , Ariful Azad
-
An Efficient Neighborhood-based Interaction Model for Recommendation on Heterogeneous Graph, KDD'20 Jiarui Jin, Jiarui Qin, Yuchen Fang, Kounianhua Du, Weinan Zhang, Yong Yu, Zheng Zhang, Alexander J. Smola
-
Learning Interaction Models of Structured Neighborhood on Heterogeneous Information Network, Jiarui Jin, Kounianhua Du, Weinan Zhang, Jiarui Qin, Yuchen Fang, Yong Yu, Zheng Zhang, Alexander J. Smola
-
Graphein - a Python Library for Geometric Deep Learning and Network Analysis on Protein Structures, Arian R. Jamasb, Pietro Lió, Tom L. Blundell
-
Graph Policy Gradients for Large Scale Robot Control, Arbaaz Khan, Ekaterina Tolstaya, Alejandro Ribeiro, Vijay Kumar
-
Heterogeneous Molecular Graph Neural Networks for Predicting Molecule Properties, Zeren Shui, George Karypis
-
Could Graph Neural Networks Learn Better Molecular Representation for Drug Discovery? A Comparison Study of Descriptor-based and Graph-based Models, Dejun Jiang, Zhenxing Wu, Chang-Yu Hsieh, Guangyong Chen, Ben Liao, Zhe Wang, Chao Shen, Dongsheng Cao, Jian Wu, Tingjun Hou
-
Principal Neighbourhood Aggregation for Graph Nets, Gabriele Corso, Luca Cavalleri, Dominique Beaini, Pietro Liò, Petar Veličković
-
Collective Multi-type Entity Alignment Between Knowledge Graphs, Qi Zhu, Hao Wei, Bunyamin Sisman, Da Zheng, Christos Faloutsos, Xin Luna Dong, Jiawei Han
-
Graph Representation Forecasting of Patient's Medical Conditions: towards A Digital Twin, Pietro Barbiero, Ramon Viñas Torné, Pietro Lió
-
Relational Graph Learning on Visual and Kinematics Embeddings for Accurate Gesture Recognition in Robotic Surgery, Yong-Hao Long, Jie-Ying Wu, Bo Lu, Yue-Ming Jin, Mathias Unberath, Yun-Hui Liu, Pheng-Ann Heng and Qi Dou
-
Dark Reciprocal-Rank: Boosting Graph-Convolutional Self-Localization Network via Teacher-to-student Knowledge Transfer, Takeda Koji, Tanaka Kanji
-
Graph InfoClust: Leveraging Cluster-Level Node Information For Unsupervised Graph Representation Learning, Costas Mavromatis, George Karypis
-
GraphSeam: Supervised Graph Learning Framework for Semantic UV Mapping, Fatemeh Teimury, Bruno Roy, Juan Sebastian Casallas, David macdonald, Mark Coates
-
Comprehensive Study on Molecular Supervised Learning with Graph Neural Networks, Doyeong Hwang, Soojung Yang, Yongchan Kwon, Kyung Hoon Lee, Grace Lee, Hanseok Jo, Seyeol Yoon, and Seongok Ryu
-
A graph auto-encoder model for miRNA-disease associations prediction, Zhengwei Li, Jiashu Li, Ru Nie, Zhu-Hong You, Wenzheng Bao
-
Graph convolutional regression of cardiac depolarization from sparse endocardial maps, STACOM 2020 workshop, Felix Meister, Tiziano Passerini, Chloé Audigier, Èric Lluch, Viorel Mihalef, Hiroshi Ashikaga, Andreas Maier, Henry Halperin, Tommaso Mansi
-
AttnIO: Knowledge Graph Exploration with In-and-Out Attention Flow for Knowledge-Grounded Dialogue, EMNLP'20, Jaehun Jung, Bokyung Son, Sungwon Lyu
-
Learning from Non-Binary Constituency Trees via Tensor Decomposition, COLING'20, Daniele Castellana, Davide Bacciu
-
Inducing Alignment Structure with Gated Graph Attention Networks for Sentence Matching, Peng Cui, Le Hu, Yuanchao Liu
-
Enhancing Extractive Text Summarization with Topic-Aware Graph Neural Networks, COLING'20, Peng Cui, Le Hu, Yuanchao Liu
-
Double Graph Based Reasoning for Document-level Relation Extraction, EMNLP'20, Shuang Zeng, Runxin Xu, Baobao Chang, Lei Li
-
Systematic Generalization on gSCAN with Language Conditioned Embedding, AACL-IJCNLP'20, Tong Gao, Qi Huang, Raymond J. Mooney
-
Automatic selection of clustering algorithms using supervised graph embedding, Noy Cohen-Shapira, Lior Rokach
-
Improving Learning to Branch via Reinforcement Learning, Haoran Sun, Wenbo Chen, Hui Li, Le Song
-
A Practical Guide to Graph Neural Networks, Isaac Ronald Ward, Jack Joyner, Casey Lickfold, Stash Rowe, Yulan Guo, Mohammed Bennamoun, code
-
APAN: Asynchronous Propagation Attention Network for Real-time Temporal Graph Embedding, SIGMOD'21, Xuhong Wang, Ding Lyu, Mengjian Li, Yang Xia, Qi Yang, Xinwen Wang, Xinguang Wang, Ping Cui, Yupu Yang, Bowen Sun, Zhenyu Guo, Junkui Li
-
Uncertainty-Matching Graph Neural Networks to Defend Against Poisoning Attacks, Uday Shankar Shanthamallu, Jayaraman J. Thiagarajan, Andreas Spanias
-
Computing Graph Neural Networks: A Survey from Algorithms to Accelerators, Sergi Abadal, Akshay Jain, Robert Guirado, Jorge López-Alonso, Eduard Alarcón
-
NHK_STRL at WNUT-2020 Task 2: GATs with Syntactic Dependencies as Edges and CTC-based Loss for Text Classification, Yuki Yasuda, Taichi Ishiwatari, Taro Miyazaki, Jun Goto
-
Relation-aware Graph Attention Networks with Relational Position Encodings for Emotion Recognition in Conversations, Taichi Ishiwatari, Yuki Yasuda, Taro Miyazaki, Jun Goto
-
PGM-Explainer: Probabilistic Graphical Model Explanations for Graph Neural Networks, Minh N. Vu, My T. Thai
-
A Generalization of Transformer Networks to Graphs, Vijay Prakash Dwivedi, Xavier Bresson
-
Discourse-Aware Neural Extractive Text Summarization, ACL'20, Jiacheng Xu, Zhe Gan, Yu Cheng, Jingjing Liu
-
Learning Robust Node Representations on Graphs, Xu Chen, Ya Zhang, Ivor Tsang, Yuangang Pan
-
Adaptive Graph Diffusion Networks with Hop-wise Attention, Chuxiong Sun, Guoshi Wu
-
The Photoswitch Dataset: A Molecular Machine Learning Benchmark for the Advancement of Synthetic Chemistry, Aditya R. Thawani, Ryan-Rhys Griffiths, Arian Jamasb, Anthony Bourached, Penelope Jones, William McCorkindale, Alexander A. Aldrick, Alpha A. Lee
-
A community-powered search of machine learning strategy space to find NMR property prediction models, Lars A. Bratholm, Will Gerrard, Brandon Anderson, Shaojie Bai, Sunghwan Choi, Lam Dang, Pavel Hanchar, Addison Howard, Guillaume Huard, Sanghoon Kim, Zico Kolter, Risi Kondor, Mordechai Kornbluth, Youhan Lee, Youngsoo Lee, Jonathan P. Mailoa, Thanh Tu Nguyen, Milos Popovic, Goran Rakocevic, Walter Reade, Wonho Song, Luka Stojanovic, Erik H. Thiede, Nebojsa Tijanic, Andres Torrubia, Devin Willmott, Craig P. Butts, David R. Glowacki, Kaggle participants
-
Adaptive Layout Decomposition with Graph Embedding Neural Networks, Wei Li, Jialu Xia, Yuzhe Ma, Jialu Li, Yibo Lin, Bei Yu, DAC'20
-
Transfer Learning with Graph Neural Networks for Optoelectronic Properties of Conjugated Oligomers, J. Chem. Phys. 154, Chee-Kong Lee, Chengqiang Lu, Yue Yu, Qiming Sun, Chang-Yu Hsieh, Shengyu Zhang, Qi Liu, and Liang Shi
-
Jet tagging in the Lund plane with graph networks, Journal of High Energy Physics 2021, Frédéric A. Dreyer and Huilin Qu
-
Global Attention Improves Graph Networks Generalization, Omri Puny, Heli Ben-Hamu, and Yaron Lipman
-
Learning over Families of Sets -- Hypergraph Representation Learning for Higher Order Tasks, SDM 2021, Balasubramaniam Srinivasan, Da Zheng, and George Karypis
-
SSFG: Stochastically Scaling Features and Gradients for Regularizing Graph Convolution Networks, Haimin Zhang, Min Xu
-
Application and evaluation of knowledge graph embeddings in biomedical data, PeerJ Computer Science 7:e341, Mona Alshahrani, Maha A. Thafar, Magbubah Essack
-
MoTSE: an interpretable task similarity estimator for small molecular property prediction tasks, bioRxiv 2021.01.13.426608, Han Li, Xinyi Zhao, Shuya Li, Fangping Wan, Dan Zhao, Jianyang Zeng
-
Reinforcement Learning For Data Poisoning on Graph Neural Networks, Jacob Dineen, A S M Ahsan-Ul Haque, Matthew Bielskas
-
Generalising Recursive Neural Models by Tensor Decomposition, IJCNN'20, Daniele Castellana, Davide Bacciu
-
Tensor Decompositions in Recursive Neural Networks for Tree-Structured Data, ESANN'20, Daniele Castellana, Davide Bacciu
-
Combining Self-Organizing and Graph Neural Networks for Modeling Deformable Objects in Robotic Manipulation, Frotiers in Robotics and AI, Valencia, Angel J., and Pierre Payeur
-
Joint stroke classification and text line grouping in online handwritten documents with edge pooling attention networks, Pattern Recognition, Jun-Yu Ye, Yan-Ming Zhang, Qing Yang, Cheng-Lin Liu
-
Toward Accurate Predictions of Atomic Properties via Quantum Mechanics Descriptors Augmented Graph Convolutional Neural Network: Application of This Novel Approach in NMR Chemical Shifts Predictions, The Journal of Physical Chemistry Letters, Peng Gao, Jie Zhang, Yuzhu Sun, and Jianguo Yu
-
A Graph Neural Network to Model User Comfort in Robot Navigation, Pilar Bachiller, Daniel Rodriguez-Criado, Ronit R. Jorvekar, Pablo Bustos, Diego R. Faria, Luis J. Manso
-
Chemistry-informed Macromolecule Graph Representation for Similarity Computation and Supervised Learning, Somesh Mohapatra, Joyce An, Rafael Gómez-Bombarelli
DGL should work on
- all Linux distributions no earlier than Ubuntu 16.04
- macOS X
- Windows 10
DGL requires Python 3.6 or later.
Right now, DGL works on PyTorch 1.5.0+, MXNet 1.6+, and TensorFlow 2.3+.
conda install -c dglteam dgl # cpu version
conda install -c dglteam dgl-cuda9.2 # CUDA 9.2
conda install -c dglteam dgl-cuda10.1 # CUDA 10.1
conda install -c dglteam dgl-cuda10.2 # CUDA 10.2
conda install -c dglteam dgl-cuda11.0 # CUDA 11.0
conda install -c dglteam dgl-cuda11.1 # CUDA 11.1
| Latest Nightly Build Version | Stable Version | |
|---|---|---|
| CPU | pip install --pre dgl |
pip install dgl |
| CUDA 9.2 | pip install --pre dgl-cu92 |
pip install dgl-cu92 |
| CUDA 10.1 | pip install --pre dgl-cu101 |
pip install dgl-cu101 |
| CUDA 10.2 | pip install --pre dgl-cu102 |
pip install dgl-cu102 |
| CUDA 11.0 | pip install --pre dgl-cu110 |
pip install dgl-cu110 |
| CUDA 11.1 | pip install --pre dgl-cu111 |
pip install dgl-cu111 |
Refer to the guide here.
| Releases | Date | Features |
|---|---|---|
| v0.4.3 | 03/31/2020 | - TensorFlow support - DGL-KE - DGL-LifeSci - Heterograph sampling APIs (experimental) |
| v0.4.2 | 01/24/2020 | - Heterograph support - TensorFlow support (experimental) - MXNet GNN modules |
| v0.3.1 | 08/23/2019 | - APIs for GNN modules - Model zoo (DGL-Chem) - New installation |
| v0.2 | 03/09/2019 | - Graph sampling APIs - Speed improvement |
| v0.1 | 12/07/2018 | - Basic DGL APIs - PyTorch and MXNet support - GNN model examples and tutorials |
Check out the open source book Dive into Deep Learning.
For those who are new to graph neural network, please see the basic of DGL.
For audience who are looking for more advanced, realistic, and end-to-end examples, please see model tutorials.
Please let us know if you encounter a bug or have any suggestions by filing an issue.
We welcome all contributions from bug fixes to new features and extensions.
We expect all contributions discussed in the issue tracker and going through PRs. Please refer to our contribution guide.
If you use DGL in a scientific publication, we would appreciate citations to the following paper:
@article{wang2019dgl,
title={Deep Graph Library: A Graph-Centric, Highly-Performant Package for Graph Neural Networks},
author={Minjie Wang and Da Zheng and Zihao Ye and Quan Gan and Mufei Li and Xiang Song and Jinjing Zhou and Chao Ma and Lingfan Yu and Yu Gai and Tianjun Xiao and Tong He and George Karypis and Jinyang Li and Zheng Zhang},
year={2019},
journal={arXiv preprint arXiv:1909.01315}
}
DGL is developed and maintained by NYU, NYU Shanghai, AWS Shanghai AI Lab, and AWS MXNet Science Team.
DGL uses Apache License 2.0.




