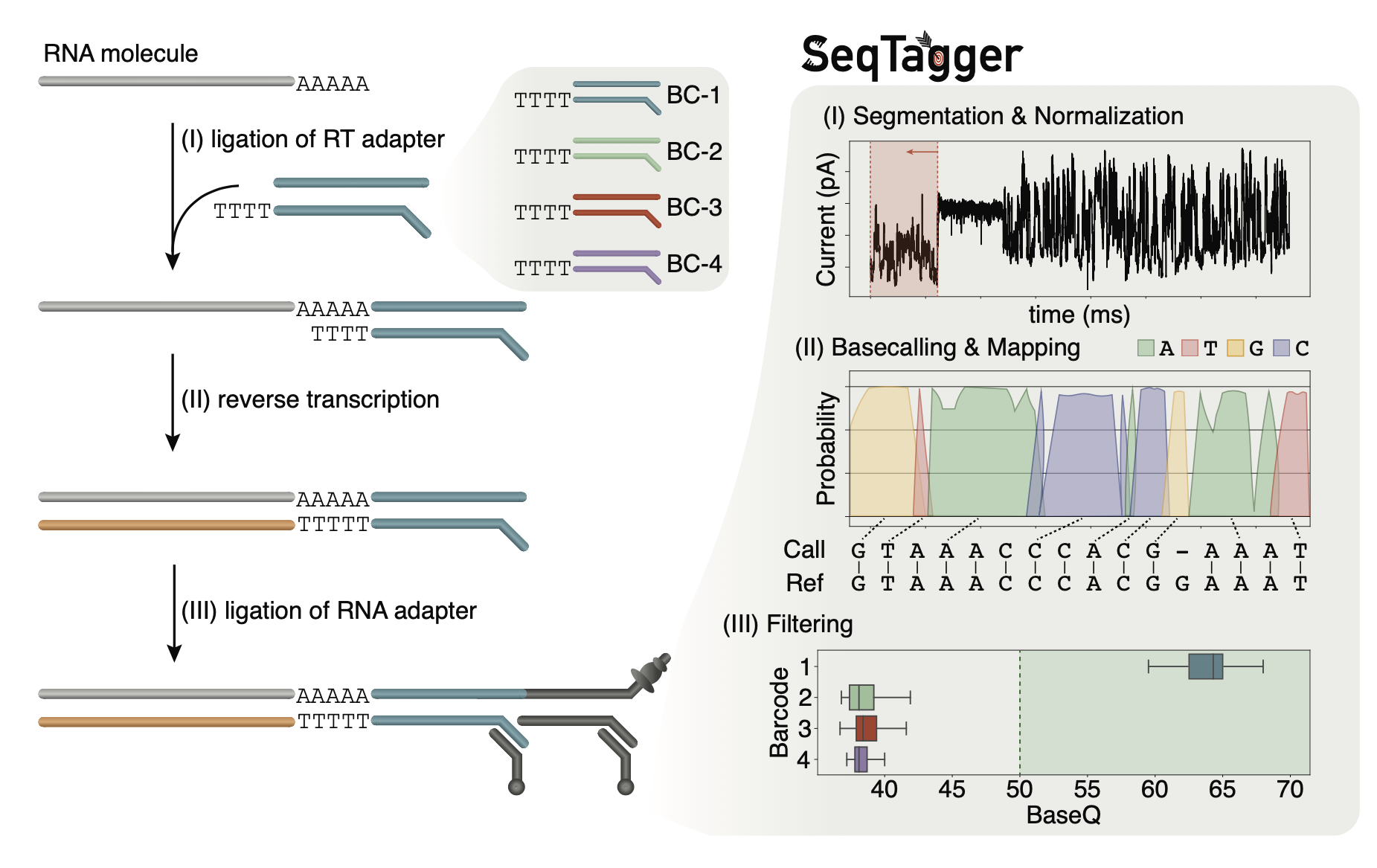
It's a super-fast and accurate demultiplexing algorithm for direct RNA nanopore sequencing datasets. Supporting both RNA002 and RNA004 kits,and both fast5 and pod5 files.
The workflow follows the standard direct RNA sequencing library preparation protocol in which default RT adapters are exchanged for barcode-containg RT adapters. SeqTagger then basecalls the DNA barcode from the direct RNA sequencing data using custom basecalling models. Finally, basecalled barcodes are aligned against the reference sequences for all barcodes and low confidence predicitions removed in a filtering step.
Currently, SeqTagger supports the following models and barcodes:
| Chemistry | Number of barcodes | SeqTagger Model | Barcode Sequences |
|---|---|---|---|
| RNA002 | 4 | b04_RNA002 | b04_RNA002_barcodes |
| RNA002 | 96 | b96_RNA002 | b96_RNA002_barcodes |
| RNA004 | 4 | b04_RNA004 | b04_RNA004_barcodes |
Please note: These models do not work well on Nano-tRNAseq libraries. You can find pre-trained tRNA demultiplexing models here.
Yes, as long as the RNA molecule has a poly(A) tail (e.g. mRNAs, lncRNAs, etc.) or you have in vitro polyadenylated the sample prior to sequencing.
Please note: Nano-tRNAseq libraries do not have standard poly(A) RNA tails, and thus should not be used with the models listed above. You can find SeqTagger Dockerfiles with pre-trained tRNA demultiplexing models here (also available for RNA004 chemistries).
Download test data for both RNA002 and RNA004:
mkdir -p seqtagger; cd seqtagger
wget https://public-docs.crg.es/enovoa/public/seqtagger/test_data/ \
-q --show-progress -r -c -nc -np -nH --cut-dirs=3 --reject="index.html*"It's handy to define an alias prior to using seqtagger:
alias seqtagger="docker run --gpus all -u $UID:$GID -v `pwd`:/data lpryszcz/seqtagger"This will bind your current directory to /data in the docker container.
Then, running it is as easy as:
seqtagger mRNA -k models/b04_RNA004 -r -i /data/test_data/RNA004 -o /data/demuxNote, you can provide multiple input directories with fast5/pod5 files after -i.
Results will be saved in tab-delimited files (gzip-compressed):
demux/RNA004.demux.tsv.gz
In addition, boxplots of per-barcode quality will be saved in corresponding directory
ending with .boxplot.pdf.
Please note: You can now also run SeqTagger through the MasterOfPores3 nextflow workflow.
If you wish to split Fast5 file(s) by barcode, execute:
seqtagger fast5_split_by_barcode.py -b 50 -i /data/demux/RNA004.demux.tsv.gz \
-f /data/test_data/RNA004 -o /data/demux/RNA004 Where -b specifies the baseQ cut-off. This will generate one output folder for each barcode named as
demux/RNA004/bc_?/reads_*.fast5 where ? represents the barcode number.
We don't provide FastQ example in the test_data. If you wish to split FastQ file(s) by barcode:
# first concatenate all FastQ file into one
cat guppy/run1/*.fastq.gz > guppy/run1.fastq.gz
# and split reads using baseQ cut-off of 50
seqtagger fastq_split_by_barcode.py -b 50 -o /data/demux/run1 -i /data/demux/run1.demux.tsv.gz -f /data/guppy/run1.fastq.gzThis will save one FastQ file for each barcode named as
demux/run1.demux.bc_?.fq.gz where ? represents the barcode number.
We don't provide BAM example in the test_data. If you wish to split BAM file(s) by barcode:
seqtagger bam_split_by_barcode.py -i /data/demux/run1.demux.tsv.gz -f /data/run1.mapped.bam -o /data/run1.mappedThis will save one BAM file for each barcode named as
run1.mapped.bc_?.bam where ? represents the barcode number.
You'll need CUDA-compatible (Nvidia) GPU and CUDA v10 or newer installed in your system.
Additionally, you'll need to install docker and NVIDIA Container Toolkit.
Versions tested:
| Software | Version |
|---|---|
| CUDA | 10, 11, 12 |
| Docker | 25+ |
| Nvidia Container Toolkit | 1.14 |
This project is licensed under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC-ND 4.0), available here, with the exception of the bonito module, which retains its original license. The full text of the licenses, including modified code, can be found in the bonito directory.
- ONT 1.0:
bonito- Licensed under the Oxford Nanopore Technologies Public License 1.0. Full license text available at ONT 1.0 License.
- MPL 2.0:
pod5,ont_fast5_api- Licensed under the Mozilla Public License 2.0. Full license text available at MPL 2.0 License.
- BSD 3-Clause:
pandas,seaborn,joblib,- Licensed under the BSD 3-Clause License. Full license text available at BSD 3-Clause License.
- MIT:
mappy,pysam,numpy- Licensed under the MIT License. Full license text available at MIT License.
- OTHER:
pytorch,numpy- Full license text for
pytorchis available at pytorch License. - Full license text for
numpyis available at numpy License.
- Full license text for
Please ensure compliance with each license's terms and conditions.
LPP, GD and EMN have filed patent applications (EP24382340 and EP24383144) based on this work at the European Patent Office.
If you found this work helpful, please cite:
Pryszcz LP*, Diensthuber G*, Llovera L, Medina R, Delgado-Tejedor A, Cozzuto L, Ponomarenko J and Novoa EM#. SeqTagger, a rapid and accurate tool to demultiplex direct RNA nanopore sequencing datasets. bioRxiv 2024 doi:[https://www.biorxiv.org/content/10.1101/2024.10.29.620808]