Table of Contents
This repository contains resources and a walkthrough for modeling presence/absence data in North Atlantic using covariates from Brickman et. al. 2021. Files included in this repository are:
- README.md (this document): Overview and walkthrough of Brickman modeling process
- brickman_walkthrough.R: the complete R code used in the modeling walkthrough
- brickman_walkthrough_help.R: code for three helper functions that can generate predictions and plots
The Brickman dataset consists of environmental covariates for a variety of climate scenarios in the North Atlantic. Covariates were generated by downscaling low-resolution IPCC data to a much higher 1/12 degree resolution. The Brickman dataset contains high resolution datasets for present day (2015), 2055, and 2075 for RCP 45 and 85 conservation scenarios. Environmental covariates available include bathymetry, bottom stress, mixed layer depth, sea surface / sea bottom temperature and salinity, and current vectors.
Brickman coordinate bounding box:
xmin = -101.5, xmax = -24.5, ymin = 16.0, ymax = 75.2
This walkthrough provides a guide for building a presence/absence model using the Brickman dataset. By training a model with present-day covariates, we can generate high resolution projections for the four available future climate situations. Example projections are in the Predictions and Plots section. This code is beginner-friendly and can perform the entire modeling process for a provided dataset, but to improve and modify the model or predictions more coding experience may be required.
- This code is for building a model that takes month as a covariate. Predicted output is on a monthly basis. Building an annual model is possible, but requires some changes to Brickman variable extraction.
- All code must be run within Bigelow’s ecocast server to function. To
run outside of ecocast, the Brickman datasets must be downloaded and
have their paths specified within
brickmanpackage methods. - This walkthrough makes use of the base R pipe operator,
|>. - Questions? Comments? Contact me by email at [email protected]
Required Packages
# for modeling and loading packages such as dplyr, purrr, and ggplot2
library(tidymodels)
# for handling Brickman data
library(stars)
library(brickman)
# for plotting
library(viridis)* The brickman package was written by Ben Tupper and is available on
ecocast and GitHub.
set.seed(607)To begin, choose your desired presence/absense data and assign it to
pa_data. The data should be an sf object (crs 4326) and have the
following columns:
PRESENCE: factor, either 0 or 1MONTH: numeric, 1 through 12
Make sure that all datapoints in pa_data are within the Brickman
coordinate bounding box (see Overview). To convert a
dataframe to sf, use
st_as_sf().
For the walkthrough, we will be using C. Finmarchicus data collected from the Ecomon survey. We will consider C. Finmarchicus present if there are more than 10,000 individuals per square meter.
library(ecomon) # not needed if not working with ecomon data
# substitute your own dataset in here
pa_data <- ecomon::read_staged(species = "calfin", form = "sf") |>
transmute(PRESENCE = (total_m2 > 10000) |> as.numeric() |> as.factor(),
MONTH = lubridate::month(date))
pa_data## Simple feature collection with 24387 features and 2 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: -75.9283 ymin: 35.2033 xmax: -65.2483 ymax: 44.77
## Geodetic CRS: WGS 84
## # A tibble: 24,387 × 3
## PRESENCE MONTH geometry
## * <fct> <dbl> <POINT [°]>
## 1 0 2 (-70.6667 41.1833)
## 2 0 2 (-70.6667 41.0167)
## 3 0 2 (-71.5 41)
## 4 0 2 (-71.0167 40.5167)
## 5 0 2 (-71.5 40.2667)
## 6 0 2 (-71.75 40.15)
## 7 0 2 (-69.9667 40.2333)
## 8 0 2 (-70 40.0167)
## 9 0 2 (-69 40.25)
## 10 0 2 (-69 40.5)
## # … with 24,377 more rows
Next, define VARS, the list of Brickman environmental covariates to
include in the modelling process. A complete list of available
covariates is below: subset this as you desire.
Bathy_depth: bathymetryXbtm: bottom stressMLD: mixed layer depthSbtm,Tbtm: sea bottom salinity, temperatureSSS,SST: sea surface salinity, temperatureU,V: horizontal and vertical current velocity
# COVARIATES
VARS <- c("Bathy_depth", "Xbtm", "MLD", "Sbtm", "SSS", "SST", "Tbtm", "U", "V")Finally, use the brickman::extract_points() method to match pa_data
to present-day Brickman covariates and create the input dataset for the
model.
pa_datais matched to Brickman covariates by pairing each presence/absence location with the geographically closest Brickman datapoint.
# pairing brickman present data to presence data
model_data <- brickman::extract_points(brickman::compose_filename("PRESENT"),
vars = VARS,
pts = pa_data,
complete = TRUE,
simplify_names = TRUE) |>
# binding the original dataset and selecting needed columns
bind_cols(pa_data) |>
select(lat, lon, PRESENCE, MONTH, all_of(VARS)) |>
rowwise() |>
# monthly variables are returned as a list with 12 values - must extract correct month
mutate_at(VARS[!VARS == "Bathy_depth"], ~.x[[MONTH]]) |>
ungroup() |>
# converting month to a factor
# remove this line to treat month as continuous ~ requires changes to get_predictions()
mutate(MONTH = as.factor(MONTH))
# input dataset for workflow
model_data## # A tibble: 24,387 × 13
## lat lon PRESENCE MONTH Bathy_depth Xbtm MLD Sbtm SSS SST Tbtm
## <dbl> <dbl> <fct> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 41.2 -70.6 0 2 36.6 0.00244 22.7 31.0 30.9 2.87 3.12
## 2 41.0 -70.6 0 2 45.1 0.00249 35.8 31.1 31.0 3.20 3.33
## 3 41.0 -71.5 0 2 43.9 0.00460 26.8 31.4 31.2 3.94 4.46
## 4 40.5 -71.0 0 2 76.6 0.00380 41.2 32.5 31.3 4.23 6.95
## 5 40.3 -71.5 0 2 85.1 0.00733 35.3 33.4 31.4 4.54 8.73
## 6 40.1 -71.8 0 2 83.4 0.00531 33.9 33.6 31.4 4.69 9.09
## 7 40.2 -69.9 0 2 93.0 0.00169 30.0 33.8 31.3 4.22 8.74
## 8 40.0 -70.0 0 2 234. 0.000930 23.2 35.0 31.5 4.79 8.35
## 9 40.3 -69.0 0 2 113. 0.00162 22.9 34.5 31.4 4.47 9.36
## 10 40.5 -69.0 0 2 74.7 0.00515 30.2 32.9 31.2 4.05 6.95
## # … with 24,377 more rows, and 2 more variables: U <dbl>, V <dbl>
Next, use Tidymodels to build a workflow object modeling
model_data. An example workflow is below, but feel free to change
recipes, models, or splitting techniques as desired.
- The example recipe converts
UandVto a singleVelattribute representing overall current velocity. - For more information on modeling with
Tidymodels, check out myTidymodels Tutorial.
# performing the initial testing/training split
# training data will be used to train the model, and the testing data is used
# to assess model performance
data_split <- initial_split(model_data, prop = 3/4, strata = PRESENCE)
training_data <- training(data_split)
testing_data <- testing(data_split)
# the recipe contains the formula for the model and data preprocessing steps
# note that the recipe passed in is modified by update_role() and step_mutate()
recipe <- recipe(PRESENCE ~ ., data = training_data) |>
update_role(lat, lon, U, V, new_role = "ID") |>
step_mutate(Vel = sqrt(U^2 + V^2), role = "predictor") |>
step_corr(all_numeric_predictors(), threshold = .95) |>
step_zv(all_predictors()) |>
step_normalize(all_numeric_predictors())
# example model specification: random forest
model <- rand_forest(trees = 15) |>
set_engine("ranger") |>
set_mode("classification")
# workflow: bundling preprocessing and model together
workflow <- workflow() |>
add_recipe(recipe) |>
add_model(model) |>
fit(training_data)
workflow## ══ Workflow [trained] ══════════════════════════════════════════════════════════
## Preprocessor: Recipe
## Model: rand_forest()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## 4 Recipe Steps
##
## • step_mutate()
## • step_corr()
## • step_zv()
## • step_normalize()
##
## ── Model ───────────────────────────────────────────────────────────────────────
## Ranger result
##
## Call:
## ranger::ranger(x = maybe_data_frame(x), y = y, num.trees = ~15, num.threads = 1, verbose = FALSE, seed = sample.int(10^5, 1), probability = TRUE)
##
## Type: Probability estimation
## Number of trees: 15
## Sample size: 18289
## Number of independent variables: 8
## Mtry: 2
## Target node size: 10
## Variable importance mode: none
## Splitrule: gini
## OOB prediction error (Brier s.): 0.1320433
Evaluate the model with whatever tools you desire. Some examples of analyses are below.
# using augment to make predictions for the test dataset
test_results <- augment(workflow, testing_data)
# note .pred_class, .pred_0 and .pred_1 columns
dplyr::glimpse(test_results)## Rows: 6,098
## Columns: 16
## $ lat <dbl> 40.78967, 42.32255, 41.01825, 41.48721, 42.26971, 42.33488…
## $ lon <dbl> -69.03500, -69.94368, -66.96130, -66.98309, -66.02243, -65…
## $ PRESENCE <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 1…
## $ MONTH <fct> 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3…
## $ Bathy_depth <dbl> 70.81680, 173.86778, 74.30029, 59.65366, 236.74553, 162.35…
## $ Xbtm <dbl> 0.029910511, 0.001796889, 0.002320968, 0.011585404, 0.0158…
## $ MLD <dbl> 35.944938, 41.321999, 23.291612, 31.152545, 22.944687, 19.…
## $ Sbtm <dbl> 32.34280, 34.18937, 33.38723, 32.41780, 34.84989, 34.64002…
## $ SSS <dbl> 31.23745, 31.21508, 31.12386, 30.95915, 30.75386, 30.79176…
## $ SST <dbl> 3.492243, 2.290613, 3.554846, 3.158253, 2.355116, 2.238144…
## $ Tbtm <dbl> 5.317258, 7.193180, 7.277824, 5.908505, 7.275045, 8.194640…
## $ U <dbl> -5.707603e-05, 1.230224e-03, -9.438548e-04, -8.058323e-03,…
## $ V <dbl> -0.069329586, -0.004909825, -0.006463766, -0.030135087, 0.…
## $ .pred_class <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0…
## $ .pred_0 <dbl> 0.99259259, 0.70500000, 0.99259259, 1.00000000, 0.85333333…
## $ .pred_1 <dbl> 0.007407407, 0.295000000, 0.007407407, 0.000000000, 0.1466…
Yardstickprovides methods to easily collect performance metrics.
# defining and retrieving desired metrics
pa_metrics <- yardstick::metric_set(roc_auc, sens, spec, accuracy)
pa_metrics(test_results,
truth = PRESENCE,
estimate = .pred_class,
.pred_1,
event_level = "second")## # A tibble: 4 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 sens binary 0.721
## 2 spec binary 0.873
## 3 accuracy binary 0.823
## 4 roc_auc binary 0.887
- By splitting the results table by month, we can collect a monthly performance breakdown.
# building a table that contains AUC for each month of predictions
auc_monthly <- count(test_results, MONTH) |>
bind_cols(AUC = split(test_results, test_results$MONTH) |>
lapply(function(x) roc_auc_vec(x$PRESENCE,
x$.pred_1,
event_level = "second")) |>
unlist()) |>
mutate(MONTH = as.numeric(MONTH))
# plotting the AUC by month
ggplot(data = auc_monthly,
mapping = aes(x = MONTH, y = AUC)) +
geom_line() +
geom_point() +
scale_x_continuous(name = "Month",
breaks = 1:12,
labels = c("Jan", "Feb", "Mar", "Apr",
"May", "Jun", "Jul", "Aug",
"Sep", "Oct", "Nov", "Dec")) +
scale_y_continuous(name = "AUC", limits = c(.5, 1)) +
ggtitle("AUC by Month") +
theme_classic() +
theme(panel.grid.major.y = element_line())Now, using workflow we can generate and visualize monthly predictions
for a desired Brickman climate sitation.
brickman_walkthrough_help.R
defines three helper functions to assist with this portion of the
modeling process:
get_predictions()takes a workflow and returns a list of by-month predictions for a desired climate situation.get_value_plots()creates plots presence probabilities from a prediction list.get_threshold_plots()creates plots showing how presence shifts between climate situations relative to a desired threshold.
First, generate predictions using get_predictions().
- Although we trained the model on present-day data, note that I’m still generating present predictions. The training data likely will not cover the entire extent of the Brickman dataset. By generating present predictions, we can see projections for areas outside of the training data’s bounds as well as create a point of reference for how predictions change between present and future climate situations.
- The
downsampleargument represents the desired resolution of predictions. 0 represents original resolution (1 datapoint per 1/12 square degrees), and higher values represent lower resolutions. For more information on howdownsamplescales the Brickman data, seestars::st_downsample(). Due to ecocast’s RAM limitations, this walkthrough can only supportdownsamplevalues of 1 or higher.- Original resolution (
downsample = 0) predictions and plots have large file sizes that will overwhelm ecocast’s RAM. To project at original resolution, I’d suggest reworking thebrickman_walkthrough_help.Rmethods so that each month of predictions is saved separately to file and plotting methods read in prediction data one month at a time.
- Original resolution (
source("brickman_walkthrough_help.R")
# predictions for the most extreme climate situation: RCP85 2075.
rcp85_2075 <- get_predictions(wkf = workflow, # a fitted workflow used to predict
brickman_vars = VARS, # needed brickman covariates
year = 2075, # year of predictions
scenario = "RCP85", # scenario of predictions
augment_preds = FALSE, # bind predictions to covariates?
verbose = FALSE,
downsample = 3) # resolution of predictions
# present day predictions
# Since I intend to compare RCP85 2075 to present day, the downsample value must be the same
present_preds <- get_predictions(wkf = workflow,
brickman_vars = VARS,
year = NA, # note that PRESENT is a scenario, not a year
scenario = "PRESENT",
augment_preds = FALSE,
verbose = FALSE,
downsample = 3)
# predictions for the least extreme future climate situation: RCP45 2055
# Generating these predictions at a higher resolution
rcp45_2055 <- get_predictions(wkf = workflow,
brickman_vars = VARS,
year = 2055,
scenario = "RCP45",
augment_preds = FALSE,
verbose = FALSE,
downsample = 1) # higher resolution
# list of 12 tibbles of prediction data - 1 per month
length(rcp85_2075)## [1] 12
# can index by month number or by 3 letter abbreviation of desired month
rcp45_2055[["Oct"]]## # A tibble: 199,783 × 6
## .pred_0 .pred_1 lon lat MONTH .pred_class
## <dbl> <dbl> <dbl> <dbl> <fct> <fct>
## 1 0.825 0.175 6.20 37.0 10 0
## 2 0.825 0.175 6.21 37.1 10 0
## 3 0.825 0.175 6.21 37.2 10 0
## 4 0.825 0.175 6.22 37.3 10 0
## 5 0.825 0.175 6.22 37.5 10 0
## 6 0.825 0.175 6.23 37.6 10 0
## 7 0.825 0.175 6.23 37.7 10 0
## 8 0.825 0.175 6.23 37.8 10 0
## 9 0.809 0.191 6.24 38.0 10 0
## 10 0.825 0.175 6.24 38.1 10 0
## # … with 199,773 more rows
There are three pre-defined ways to visualize prediction data:
- Raw: Plot the predicted presence probability for a climate situation. This is best for understanding where a model places presences and absences.
- Difference: Plot the difference in presence probability between two different climate situations. This is best for understanding how raw probabilities may shift over time.
- Threshold: Plot how presence probability shifts relative to a desired threshold. This is best for examining how high-presence areas shift between climate situations.
Use get_value_plots() to retrieve raw or difference plots, and use
get_threshold_plots() to retrieve threshold plots. Both of these
methods return a list of 12 ggplot objects named by month.
- If creating a difference or threshold plot, ensure that the downsample value is the same for both the original and comparison predictions.
# raw plots for RCP85 2075
raw_plots_rcp85_2075 <- get_value_plots(preds_list = rcp85_2075, # prediction data
title = "RCP85 2075 Predicted Presence Probability",
# size of points in graph - optimal value depends on crop and downsample
pt_size = .3,
xlim = NULL, # optional bounds for plot, if zooming in on an area
ylim = NULL)
# Where are the predicted presence probabilities for RCP85 2075 in October?
raw_plots_rcp85_2075[["Oct"]]# another example of raw plots, now for RCP45 2055
# This plot is cropped to the Gulf of Maine / Gulf of St. Lawrence
raw_plots_rcp45_2055 <- get_value_plots(preds_list = rcp45_2055,
title = "RCP45 2055 Predicted Presence Probability",
pt_size = .4, # note adjusted point size
xlim = c(-77.0, -42.5), # cropping
ylim = c(36.5, 56.7))
# What are the predicted presence probabilities for RCP45 2055 in May in the GoM/GSL?
raw_plots_rcp45_2055[["May"]]# difference plots for RCP85 2075 relative to present day
difference_plots <- get_value_plots(preds_list = rcp85_2075,
title = "RCP85 2075 Change in Presence Probability",
pt_size = .3,
xlim = NULL,
ylim = NULL,
# optional comparison argument indicates need for
comparison_list = present_preds)
# How have probabilities shifted between present day and RCP85 2075?
difference_plots[["Oct"]]# threshold plots for RCP85 2075, relative to present day
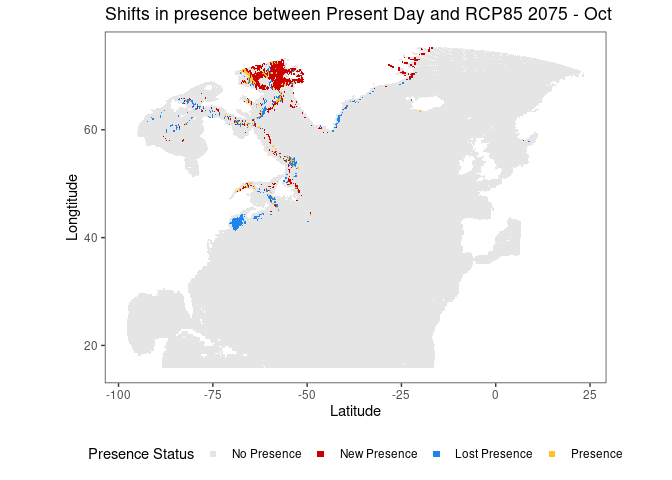
threshold_plots <- get_threshold_plots(preds_list = rcp85_2075, # target prediction data
comparison_list = present_preds, # comparison prediction data
threshold = .5, # threshold for presence
title = "Shifts in presence between Present Day and RCP85 2075",
pt_size = .3,
xlim = NULL,
ylim = NULL)
# Where are the new, lost, and maintained presence areas in RCP85 2075 relative to present day?
threshold_plots[["Oct"]]