Tool for analysis of copy number profile in WGS.
git clone https://github.com/stjude/doCNA.git
# setup a virtual environment
python -m venv docna_install
source ./docna_install/bin/activate
# install into virtual environment
pip3 install ./doCNA- Get a local copy of the config:
docna getconfig-
Edit the config to point to real SuperGood and CytoBand files. We don't recommend changing additional parameters when starting out.
-
Run doCNA to generate output files:
# replace sample with whatever your samplename is
docna analyze -i Sample.txt -c doCNA.ini -s Sample
... Lots of log info! ...
12:18:19 doCNA.WGS: INFO: Ready to report!
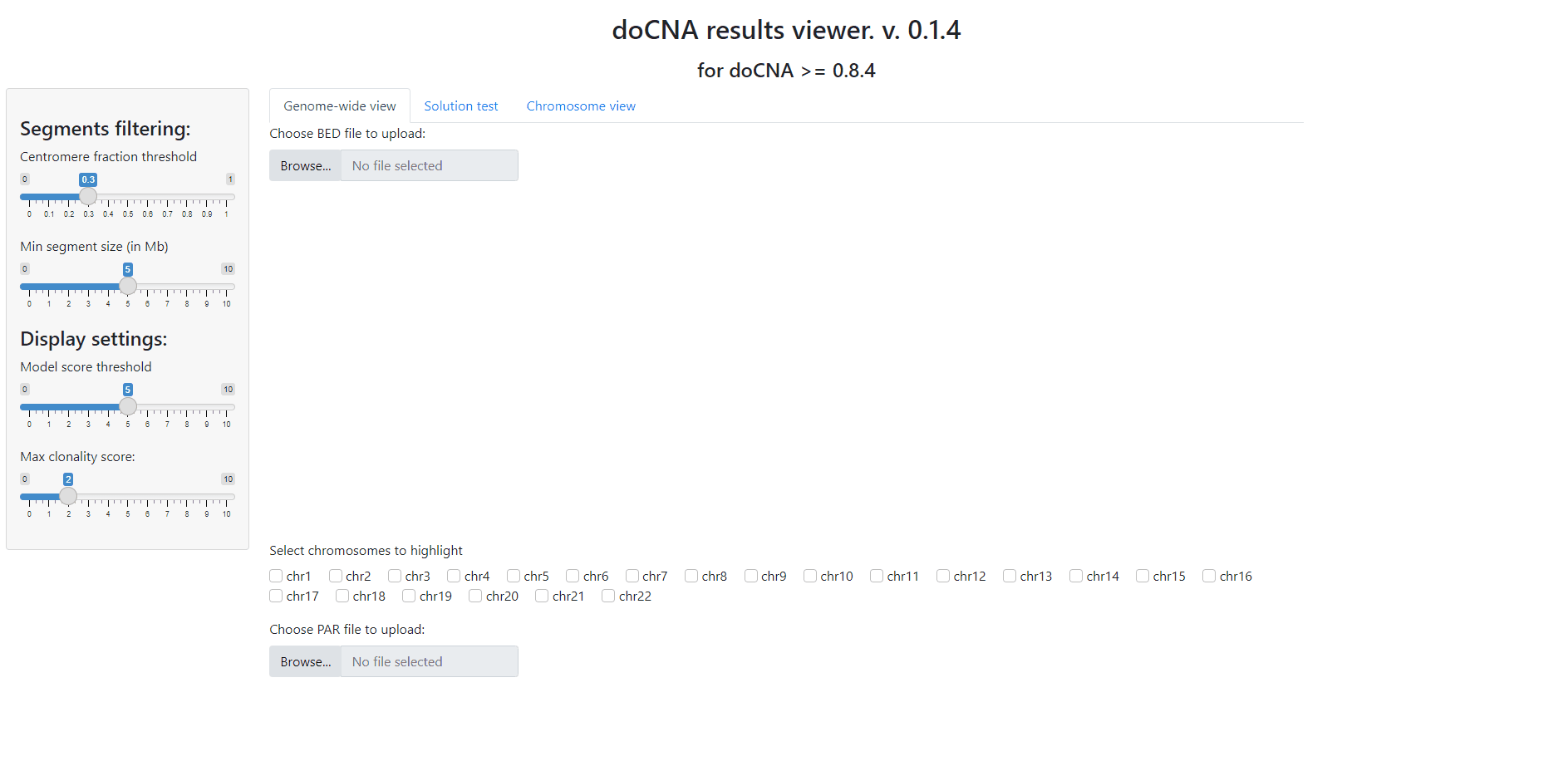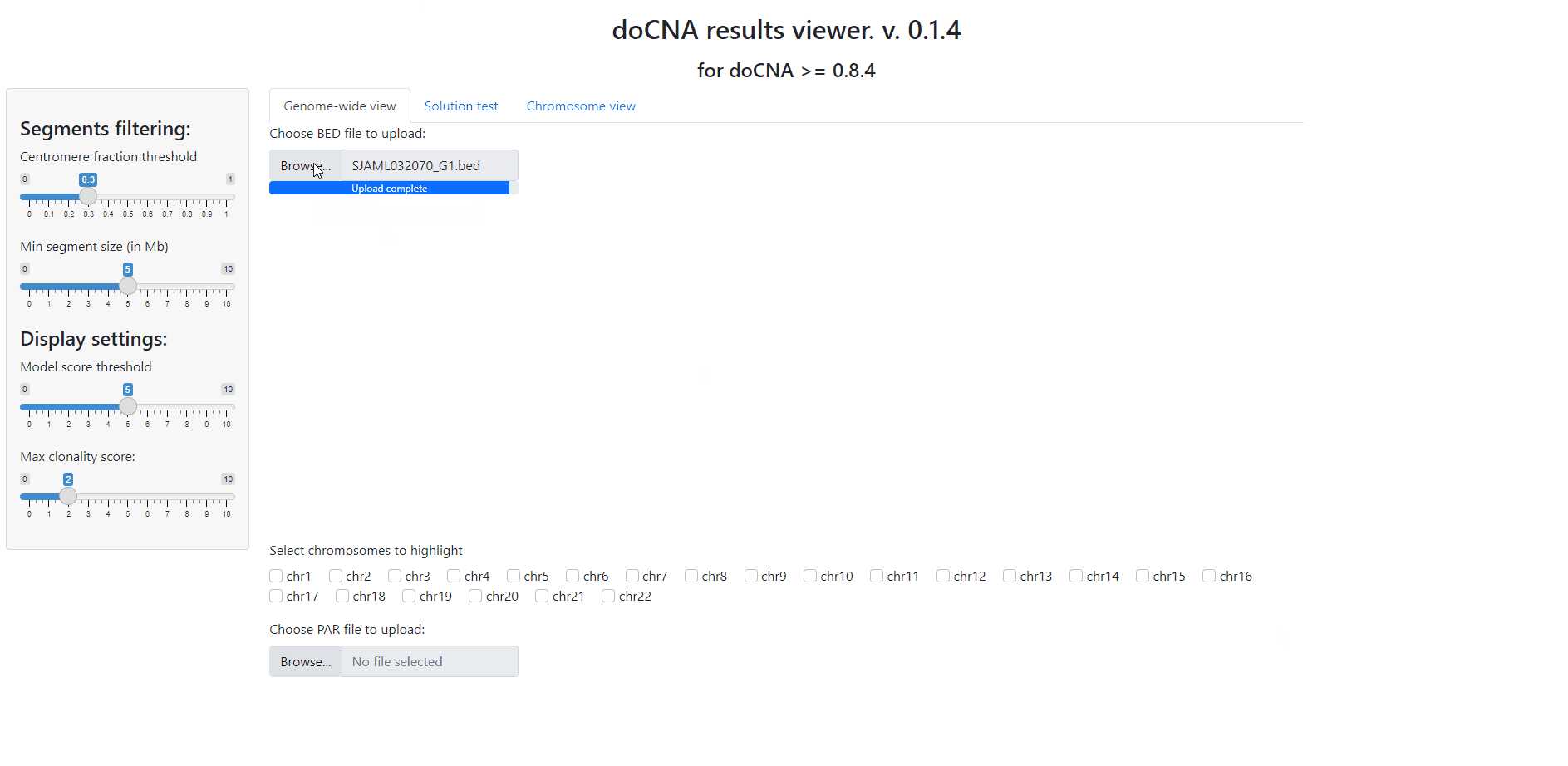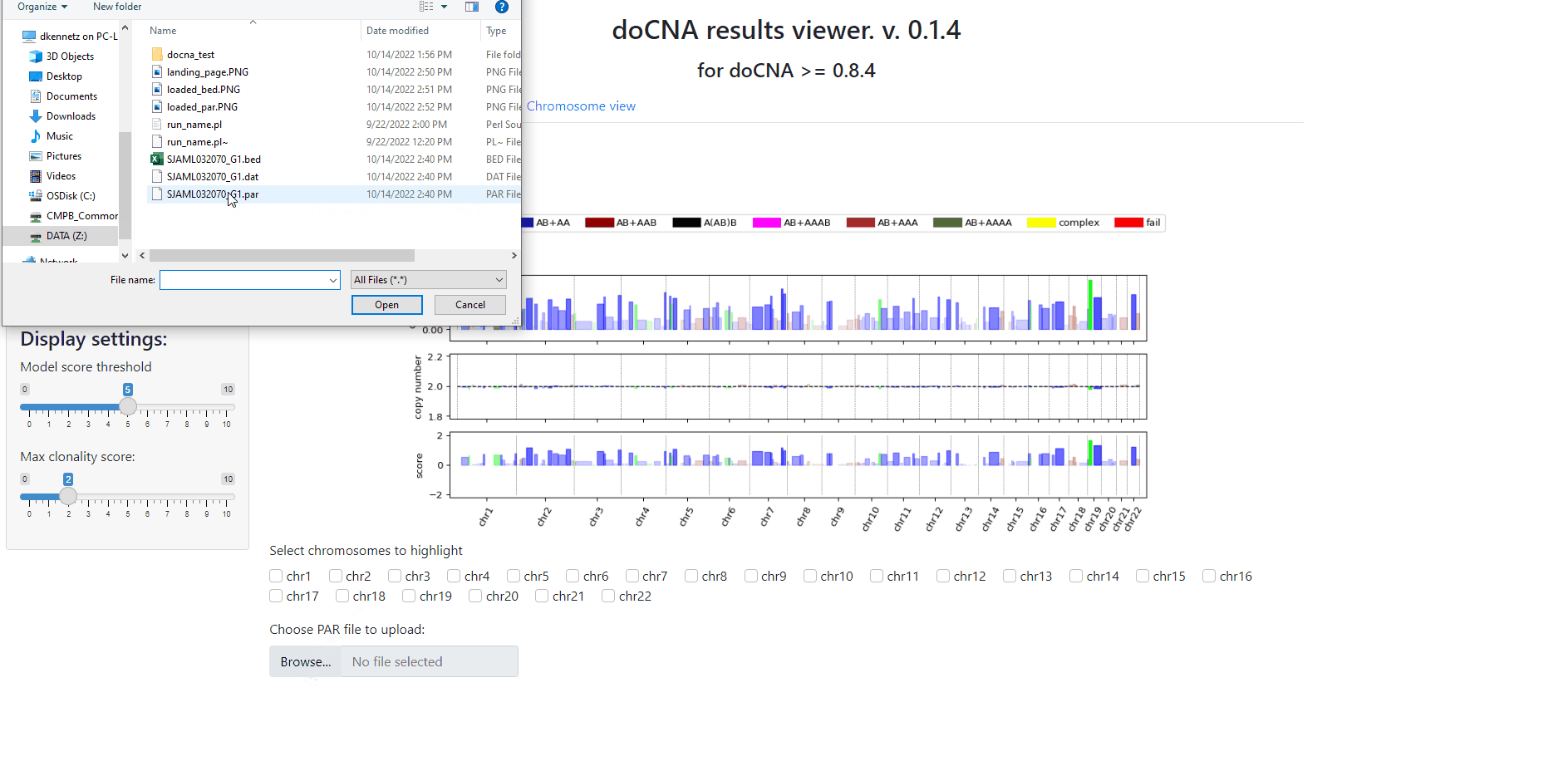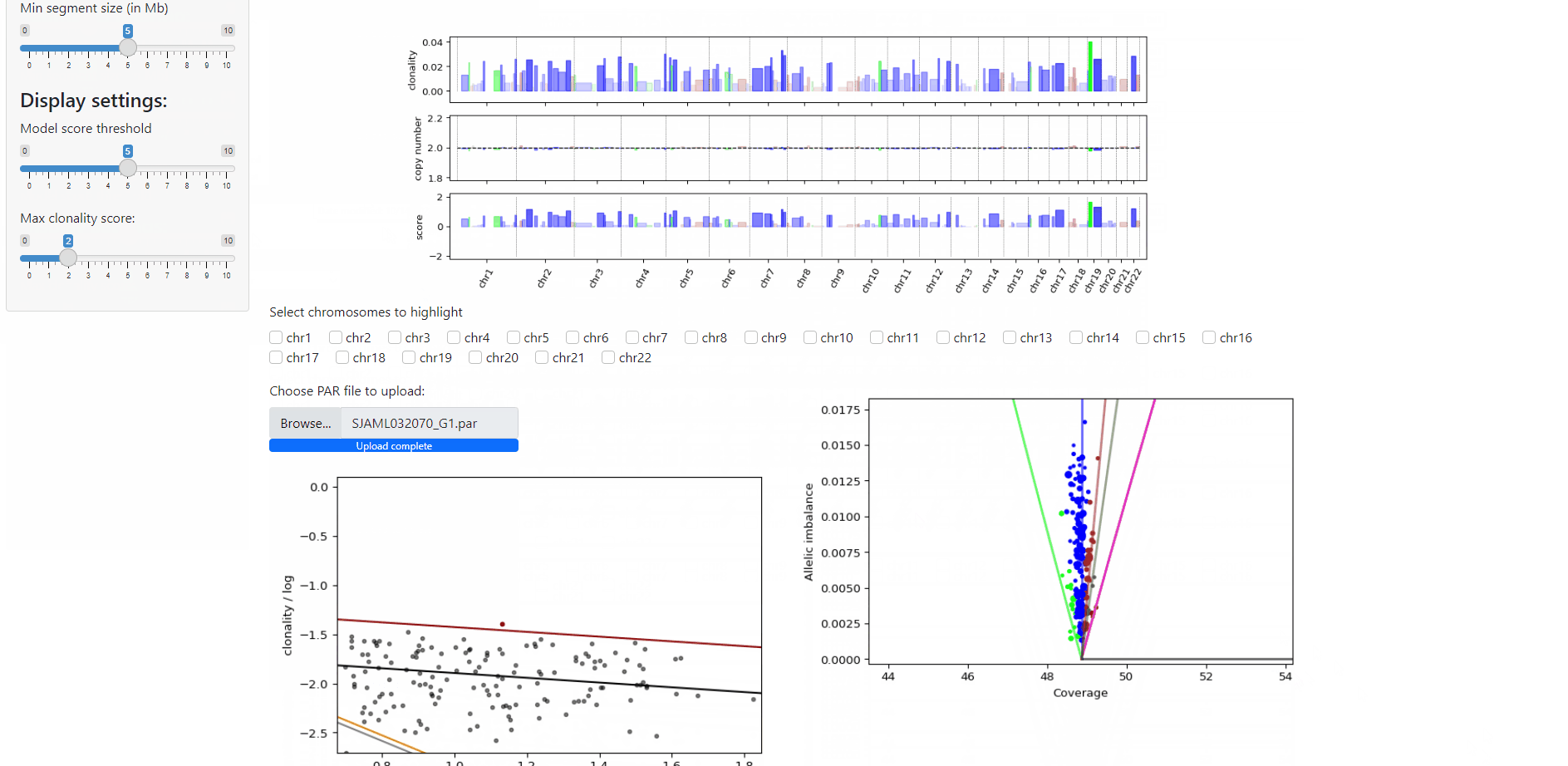
All done- Launch the viewer and visualize output:
# if you are running this on your local machine, such as a desktop or laptop:
docna viewer
# if you are running this on a remote machine, like a cluster:
docna viewer --remote
**********
Access dashboard in browser via: http://10.220.16.129:39738
**********
INFO: Started server process [11716]
INFO: Waiting for application startup.
INFO: Application startup complete.
INFO: Uvicorn running on http://10.220.16.129:39738 (Press CTRL+C to quit)