Master’s Thesis in Computational Biology
Technical University of Denmark, 2015
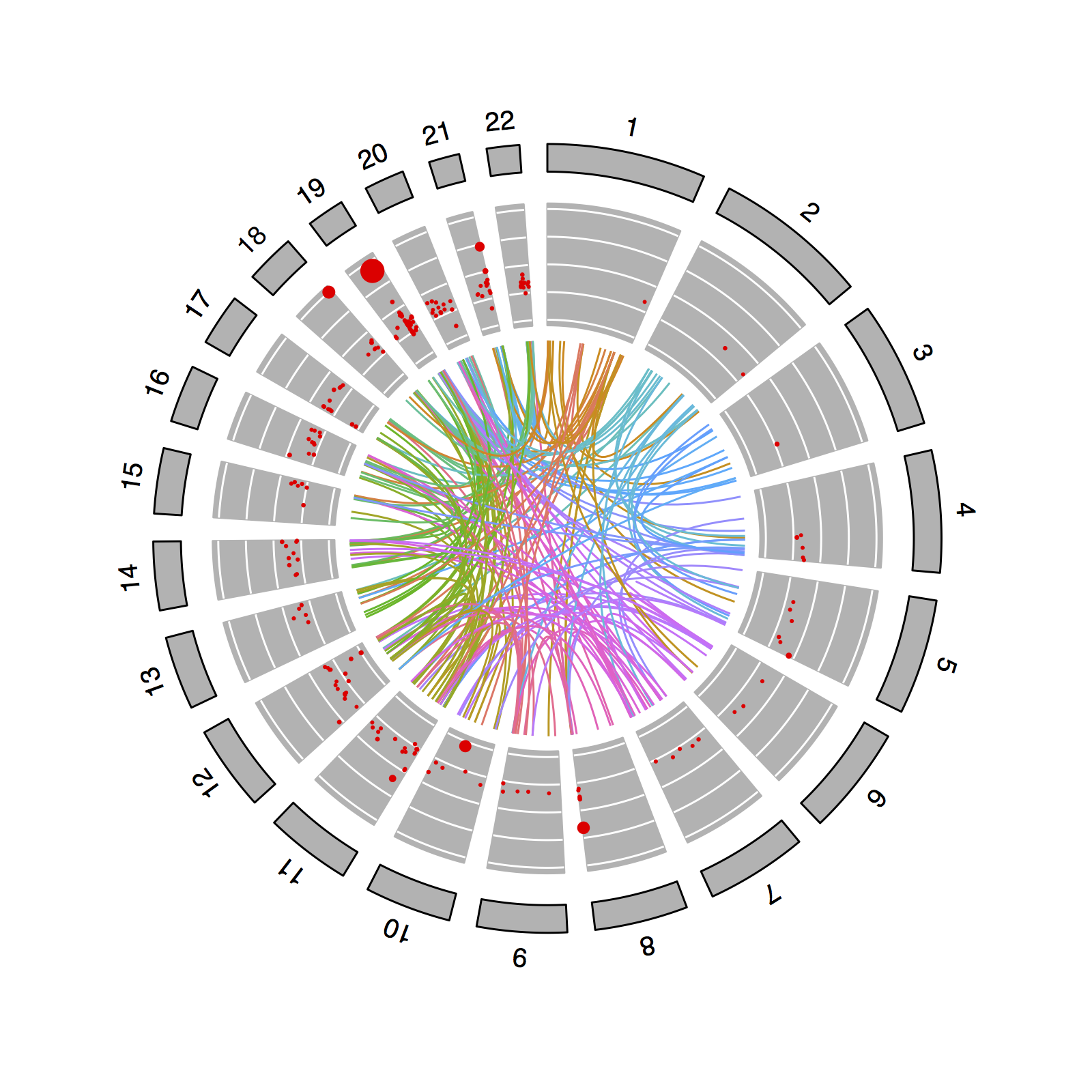
Genome-wide association studies (GWAS) successfully identified thousands of loci harbouring genetic variants associated with common human diseases and traits. GWAS has elucidated novel biological disease mechanisms, but have so far been limited to single-locus associations. Single-nucleotide polymorphisms (SNPs) are often modeled as having independent additive effects on the phenotype, neglecting any interaction effects between SNPs. Epistasis (that is, the statistical interaction between SNPs) has been demonstrated to prevail in model organisms such as yeast, but results have been limited in human studies.
Our knowledge of the mechanisms that can cause epistasis is still very incomplete. Arguably, large-scale detection of epistasis has previously been too technically challenging owing to statistical and computational issues - the combinatorial explosion of statistical tests becomes intractable when considering a genome scan of all two-loci interactions. Thus, efficient computational methods using a priori biological knowledge are needed to constrain the search space for epistasis.
Recent advances in chromosome conformation capture techniques, such as the Hi-C, have generated 3D maps of the human genome. With this a new paradigm in genome biology has emerged: genomes are organized around gene regulatory factors that govern cell identity. This has left a hitherto unexplored territory of relationships between spatial and genetic factors.
My thesis aims to uncover regulatory molecular mechanisms that can cause epistatic interactions influencing human gene expression. I hypothesize that physical prox- imity of interacting genomic regions provides a spatial scaffold to identify genetic interactions in human genotyping data. Specifically, I focus on the question whether spatial interacting genomic loci do enrich for epistasis. To the best of my knowledge, this is the first work that connects 3D genome maps and genetic data to guide the search for epistasis.
By leveraging data integration of genome-wide genetic, transcriptomic and Hi-C data, I developed a novel computational framework for performing enrichment analysis of “spatial epistasis” using a priori biological knowledge. I argue that the framework can be generalized to support more advanced statistical models of epistasis and, more importantly, alternative biological priors to drive the search for interacting SNPs.
The hypothesis was tested under different scenarios of spatial epistasis, including genome structures from multiple cell lines. For one of the cell lines tested, I identified enrichment of spatial epistasis. However, the signal was eliminated, after more thorough quality control of the data. I conclude that this study was underpowered to detect epistasis at this scale. Nevertheless, these results does not support the existence of prevailing epistasis influencing human gene expression. It remains inconclusive whether spatially interacting genomic regions are enriched for epistasis, and ultimately, there is no evidence to definitively reject or accept the spatial epistasis hypothesis.