SingleCellNet enables the classifcation of single cell RNA-Seq data across species and platforms. See BioRxiv for details.
Here, we illustrate ...
-
how to build and assess single cell classifiers
-
how to build and assess cross-species single cell classifiers
-
how to use these classifiers to quantify 'cell identity' from query scRNA-Seq data
If you want to use the bulk RNA-Seq version of CellNet, go to bulk CellNet.
In this example, we use a subset of the Tabula Muris data to train singleCellNet. To learn more about the Tabula Muris project, see the manuscript. As query data, we use scRNA-Seq of kidney cells as reported in Park et al 2018. We also provide an example of classifying human, bead enriched PBMCs (from https://www.ncbi.nlm.nih.gov/pubmed/28091601). You can download this data here:
| APPLICATION | METADATA | EXPRESSION |
|---|---|---|
| Query | metadata | expression data |
| Training | metadata | expression data |
| cross-species | human-mouse orthologs | Query (human bead-purified PBMC from 10x) |
install.packages("devtools")
library(devtools)
install_github("thomasp85/patchwork")
install_github("pcahan1/singleCellNet")
library(singleCellNet)
library(dplyr)
mydate<-utils_myDate()devtools::install_github(repo = "hhoeflin/hdf5r")
devtools::install_github(repo = "mojaveazure/loomR", ref = "develop")
library(loomR)
download.file("https://s3.amazonaws.com/cnobjects/singleCellNet/examples/sampTab_Park_MouseKidney_062118.rda", "sampTab_Park_MouseKidney_062118.rda")
download.file("https://s3.amazonaws.com/cnobjects/singleCellNet/examples/expMatrix_Park_MouseKidney_Oct_12_2018.rda", "expMatrix_Park_MouseKidney_Oct_12_2018.rda")
download.file("https://s3.amazonaws.com/cnobjects/singleCellNet/examples/expMatrix_TM_Raw_Oct_12_2018.rda", "expMatrix_TM_Raw_Oct_12_2018.rda")
download.file("https://s3.amazonaws.com/cnobjects/singleCellNet/examples/sampTab_TM_053018.rda", "sampTab_TM_053018.rda")
## For cross-species analyis:
download.file("https://s3.amazonaws.com/cnobjects/singleCellNet/examples/human_mouse_genes_Jul_24_2018.rda", "human_mouse_genes_Jul_24_2018.rda")
download.file("https://s3.amazonaws.com/cnobjects/singleCellNet/examples/6k_beadpurfied_raw.rda", "6k_beadpurfied_raw.rda")
download.file("https://s3.amazonaws.com/cnobjects/singleCellNet/examples/stDat_beads_mar22.rda", "stDat_beads_mar22.rda")
## To demonstrate how to integrate loom files to SCN
download.file("https://s3.amazonaws.com/cnobjects/singleCellNet/examples/pbmc_6k.loom", "pbmc_6k.loom")stPark<-utils_loadObject("sampTab_Park_MouseKidney_062118.rda")
expPark<-utils_loadObject("expMatrix_Park_MouseKidney_Oct_12_2018.rda")
dim(expPark)
[1] 16272 43745
genesPark<-rownames(expPark)
rm(expPark)
gc()expTMraw<-utils_loadObject("expMatrix_TM_Raw_Oct_12_2018.rda")
dim(expTMraw)
[1] 23433 24936
stTM<-utils_loadObject("sampTab_TM_053018.rda")
dim(stTM)
[1] 24936 17
stTM<-droplevels(stTM)commonGenes<-intersect(rownames(expTMraw), genesPark)
length(commonGenes)
[1] 13831
expTMraw<-expTMraw[commonGenes,]stList<-splitCommon(stTM, ncells=100, dLevel="newAnn")
stTrain<-stList[[1]]
expTrain<-expTMraw[,rownames(stTrain)]
system.time(tmpX<-weighted_down(expTrain, 1.5e3, dThresh=0.25))
user system elapsed
5.662 0.958 6.649
system.time(expTrain<-trans_prop(tmpX, 1e4))
user system elapsed
2.033 0.888 2.925 system.time(cgenes2<-findClassyGenes(expTrain, stTrain, "newAnn", topX=10))
user system elapsed
51.145 7.603 59.067
cgenesA<-cgenes2[['cgenes']]
grps<-cgenes2[['grps']]
length(cgenesA)
[1] 473
# heatmap these genes
hm_gpa_sel(expTrain, cgenesA, grps, maxPerGrp=5, toScale=T, cRow=F, cCol=F,font=4)expT<-as.matrix(expTrain[cgenesA,])
dim(expT)
[1] 473 3036
system.time(xpairs<-ptGetTop(expT, grps, topX=25, sliceSize=5000))
user system elapsed
1902.520 1189.397 803.201
length(xpairs)
[1] 797system.time(pdTrain<-query_transform(expT[cgenesA, ], xpairs))
user system elapsed
0.179 0.031 0.212
dim(pdTrain)
[1] 797 3036
system.time(rf_tspAll<-sc_makeClassifier(pdTrain[xpairs,], genes=xpairs, groups=grps, nRand=100, ntrees=1000))
user system elapsed
393.570 1.112 395.620 stTest<-stList[[2]]
system.time(expQtransAll<-query_transform(expTMraw[cgenesA,rownames(stTest)], xpairs))
user system elapsed
16.479 1.578 18.359
nrand<-100
system.time(classRes_val_all<-rf_classPredict(rf_tspAll, expQtransAll, numRand=nrand))
user system elapsed
30.808 1.379 32.210 tm_heldoutassessment <- assess_comm(ct_scores = classRes_val_all, stTrain = stTrain, stQuery = stTest, dLevelSID = "cell", classTrain = "newAnn", classQuery = "newAnn")
plot_PRs(tm_heldoutassessment)plot_metrics(tm_heldoutassessment)sla<-as.vector(stTest$newAnn)
names(sla)<-rownames(stTest)
slaRand<-rep("rand", nrand)
names(slaRand)<-paste("rand_", 1:nrand, sep='')
sla<-append(sla, slaRand)
sc_hmClass(classRes_val_all, sla, max=300, isBig=TRUE)plot_attr(classRes_val_all, stTest, nrand=nrand, dLevel="newAnn", sid="cell")system.time(umPrep<-prep_umap_class(classRes_val_all, stTest, nrand=nrand, dLevel="newAnn", sid="cell", topPC=5))
user system elapsed
130.096 2.735 133.189
plot_umap(umPrep)expPark<-utils_loadObject("expMatrix_Park_MouseKidney_Oct_12_2018.rda")
system.time(kidTransAll<-query_transform(expPark[cgenesA,], xpairs))
user system elapsed
23.536 1.721 25.338
nqRand<-100
system.time(crParkall<-rf_classPredict(rf_tspAll, kidTransAll, numRand=nqRand))
user system elapsed
60.601 2.898 63.654
sgrp<-as.vector(stPark$description1)
names(sgrp)<-rownames(stPark)
grpRand<-rep("rand", nqRand)
names(grpRand)<-paste("rand_", 1:nqRand, sep='')
sgrp<-append(sgrp, grpRand)
# heatmap classification result
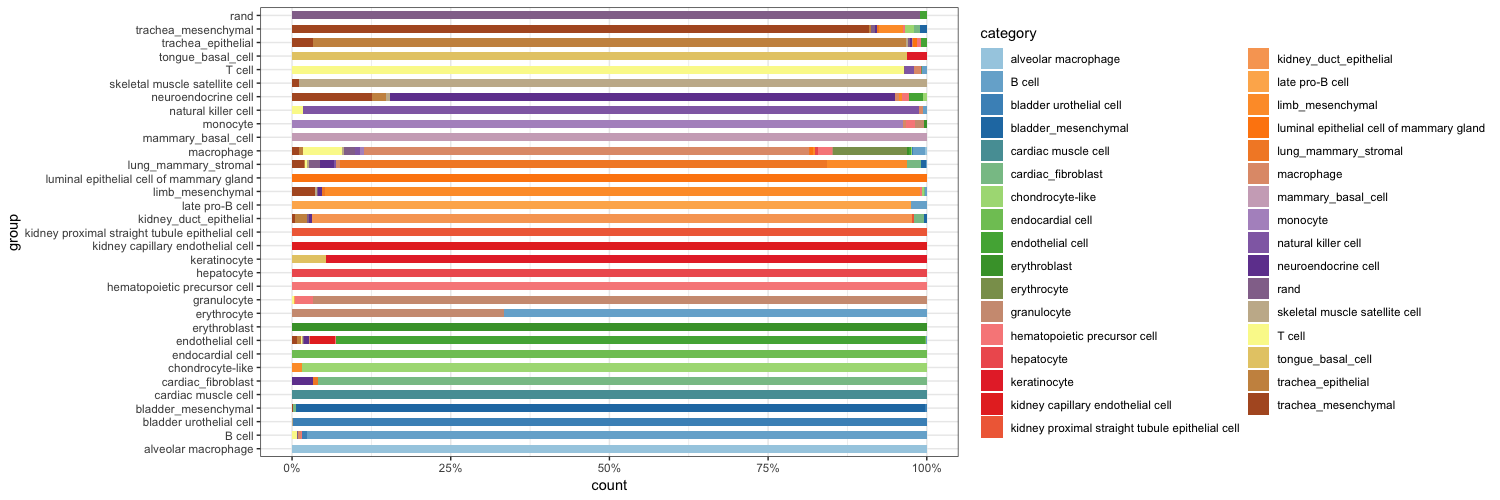
sc_hmClass(crParkall, sgrp, max=5000, isBig=TRUE, cCol=F, font=8)sc_violinClass(sampTab = stPark, classRes = crParkall, cellIDCol = "sample_name", dLevel = "description1", addRand = nrand)stKid2<-addRandToSampTab(crParkall, stPark, "description1", "sample_name")
skylineClass(crParkall, "T cell", stKid2, "description1",.25, "sample_name")stQuery<-utils_loadObject("stDat_beads_mar22.rda")
expQuery<-utils_loadObject("6k_beadpurfied_raw.rda") # use Matrix if RAM low
dim(expQuery)
[1] 32643 6000
expTMraw<-utils_loadObject("expMatrix_TM_Raw_Oct_12_2018.rda") # reload training
Load the ortholog table and convert human gene names to mouse ortholog names, and limit analysis to genes in common between the training and query data.
oTab<-utils_loadObject("human_mouse_genes_Jul_24_2018.rda")
dim(oTab)
[1] 16688 3
aa = csRenameOrth(expQuery, expTMraw, oTab)
expQuery <- aa[['expQuery']]
expTrain <- aa[['expTrain']]cts<-c("B cell", "cardiac muscle cell", "endothelial cell", "erythroblast", "granulocyte", "hematopoietic precursor cell", "late pro-B cell", "limb_mesenchymal", "macrophage", "mammary_basal_cell", "monocyte", "natural killer cell", "T cell", "trachea_epithelial", "trachea_mesenchymal")
stTM2<-filter(stTM, newAnn %in% cts)
stTM2<-droplevels(stTM2)
rownames(stTM2)<-as.vector(stTM2$cell) # filter strips rownames
expTrain<-expTrain[,rownames(stTM2)]
dim(expTrain)
[1] 14550 15161stList<-splitCommon(stTM2, ncells=100, dLevel="newAnn")
stTrain<-stList[[1]]
dim(stTrain)
[1] 1457 17
expTrain<-trans_prop(weighted_down(expTrain[,rownames(stTrain)], 1.5e3, dThresh=0.25), 1e4)
system.time(cgenes2<-findClassyGenes(expTrain, stTrain, "newAnn", topX=10))
user system elapsed
16.888 3.499 20.506
cgenesA<-cgenes2[['cgenes']]
grps<-cgenes2[['grps']]
length(cgenesA)
[1] 245
# heatmap these genes
hm_gpa_sel(expTrain, cgenesA, grps, maxPerGrp=20, toScale=T, cRow=F, cCol=F,font=4)system.time(xpairs<-ptGetTop(expTrain[cgenesA,], grps, topX=25, sliceSize=5000))
user system elapsed
185.198 142.132 163.631
length(xpairs)
[1] 374
pdTrain<-query_transform(expTrain[cgenesA, rownames(stTrain)], xpairs)
dim(pdTrain)
[1] 374 1457
nrand = 50
system.time(rf_tspAll<-sc_makeClassifier(pdTrain[xpairs,], genes=xpairs, groups=grps, nRand=nrand, ntrees=1000))
user system elapsed
18.321 0.057 18.373stTest<-stList[[2]]
system.time(expQtransAll<-query_transform(expTrain[cgenesA,rownames(stTest)], xpairs))
user system elapsed
3.055 0.375 3.489
nrand<-50
system.time(classRes_val_all<-rf_classPredict(rf_tspAll, expQtransAll, numRand=nrand))
user system elapsed
7.055 0.254 7.311 tm_heldoutassessment <- assess_comm(ct_scores = classRes_val_all, stTrain = stTrain, stQuery = stTest, dLevelSID = "cell", classTrain = "newAnn", classQuery = "newAnn")
plot_PRs(tm_heldoutassessment)plot_metrics(tm_heldoutassessment)sla<-as.vector(stTest$newAnn)
names(sla)<-rownames(stTest)
slaRand<-rep("rand", nrand)
names(slaRand)<-paste("rand_", 1:nrand, sep='')
sla<-append(sla, slaRand)
# heatmap classification result
sc_hmClass(classRes_val_all, sla, max=300, font=7, isBig=TRUE)plot_attr(classRes_val_all, stTest, nrand=nrand, dLevel="newAnn", sid="cell")system.time(umPrep<-prep_umap_class(classRes_val_all, stTest, nrand=nrand, dLevel="newAnn", sid="cell", topPC=5))
user system elapsed
79.475 2.110 82.004
plot_umap(umPrep)system.time(expQueryTrans<-query_transform(expQuery[cgenesA,], xpairs))
user system elapsed
0.308 0.371 0.743
nqRand<-50
system.time(crHS<-rf_classPredict(rf_tspAll, expQueryTrans, numRand=nqRand))
user system elapsed
3.592 0.126 3.747 stQuery$description <- as.character(stQuery$description)
stQuery[which(stQuery$description == "NK cell"), "description"] = "natural killer cell"
tm_pbmc_assessment <- assess_comm(ct_scores = crHS, stTrain = stTrain, stQuery = stQuery, classTrain = "newAnn",classQuery="description",dLevelSID="sample_name")
plot_PRs(tm_pbmc_assessment)plot_metrics(tm_pbmc_assessment)sgrp<-as.vector(stQuery$prefix)
names(sgrp)<-rownames(stQuery)
grpRand<-rep("rand", nqRand)
names(grpRand)<-paste("rand_", 1:nqRand, sep='')
sgrp<-append(sgrp, grpRand)
sc_hmClass(crHS, sgrp, max=5000, isBig=TRUE, cCol=F, font=8)Note that the macrophage category seems to be promiscuous in the mouse held out data, too.
sc_violinClass(sampTab = stQuery, classRes = crHS, cellIDCol = "sample_name", dLevel = "description")plot_attr(crHS, stQuery, nrand=nqRand, sid="sample_name", dLevel="description")system.time(umPrep_HS<-prep_umap_class(crHS, stQuery, nrand=nqRand, dLevel="description", sid="sample_name", topPC=5))
user system elapsed
26.905 1.014 27.993
plot_umap(umPrep_HS)lfile <- loadLoomExpCluster("pbmc_6k.loom", cellNameCol = "obs_names", xname = "description")
stQuery = lfile$sampTab
dim(stQuery)
[1] 6000 2
expQuery = lfile$expDat
dim(expQuery)
[1] 32643 6000
With this you can rerun the cross-species analysis and follow the exact same steps