This code was developed during the master's project 'Genetic Algorithms and Convolutional Neural Networks to Aid the Diagnosis of Vertebral Compression Fractures' of the Graduate Program in Applied Computing at the Faculty of Philosophy, Sciences and Letters of Ribeirão Preto, University of São Paulo.
- Computer-Aided Diagnosis of Vertebral Compression Fractures Using Convolutional Neural Networks and Radiomics - Journal of Digital Imaging
Current stable version: 2.0.0
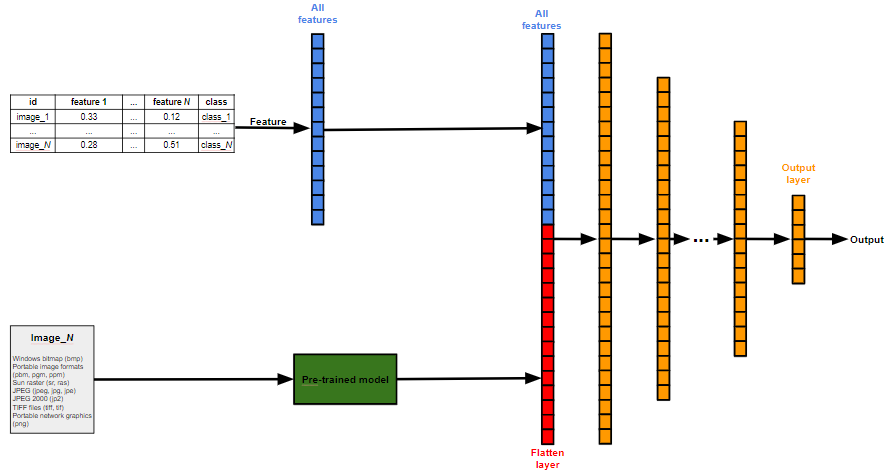
Neural Network optimized by Genetic Algorithms - (NNGA) is a library for deep model training, for data classification and segmentation. The adjustment of the model parameters can be done by genetic algorithm or can be defined by pre-trained models. For classification task, feature selection can be done by a genetic algorithm.
The NNGA works well for medical data, because with NNGA is possible training a model using both image exam result and laboratory exam /paciente information simultaneously. NNGA also preserves the image ratio, filling the background with black to maintain the actual proportion of the exam.
The model accepts two data entry sources:
-
Images
The format of the images should be:
- Windows bitmap (bmp)
- Portable image formats (pbm, pgm, ppm)
- Sun raster (sr, ras)
- JPEG (jpeg, jpg, jpe)
- JPEG 2000 (jp2)
- TIFF files (tiff, tif)
- Portable network graphics (png)
-
CSV file
2.1. The sample class should be in the column with label "class"
2.2 If the two data sources are used simultaneously, the CSV file should be a column with label "id" containing the corresponding image name (without extension)
git clone https://github.com/rafaelsdellama/nnga.git
cd nnga
pip install .The table below shows all the models that can be created:
| TASK | ARCHITECTURE | BACKBONE | FEATURE_SELECTION |
|---|---|---|---|
| Classification | MLP | GASearch | True |
| Classification | MLP | GASearch | False |
| Classification | MLP | MLP | True |
| Classification | MLP | MLP | False |
| Classification | CNN | GASearch | - |
| Classification | CNN | One of the pre-trained models: VGG16 VGG19 ResNet50 ResNet101 ResNet152 ResNet50V2 ResNet101V2 ResNet152V2 InceptionV3 InceptionResNetV2 MobileNet MobileNetV2 DenseNet121 DenseNet169 DenseNet201 NASNetMobile NASNetLarge |
- |
| Classification | CNN/MLP | GASearch | True |
| Classification | CNN/MLP | GASearch | False |
| Classification | CNN/MLP | One of the pre-trained models: VGG16 VGG19 ResNet50 ResNet101 ResNet152 ResNet50V2 ResNet101V2 ResNet152V2 InceptionV3 InceptionResNetV2 MobileNet MobileNetV2 DenseNet121 DenseNet169 DenseNet201 NASNetMobile NASNetLarge |
True |
| Classification | CNN/MLP | One of the pre-trained models: VGG16 VGG19 ResNet50 ResNet101 ResNet152 ResNet50V2 ResNet101V2 ResNet152V2 InceptionV3 InceptionResNetV2 MobileNet MobileNetV2 DenseNet121 DenseNet169 DenseNet201 NASNetMobile NASNetLarge |
False |
| Segmentation | CNN | unet | - |
- TASK: Classification
Parameters:
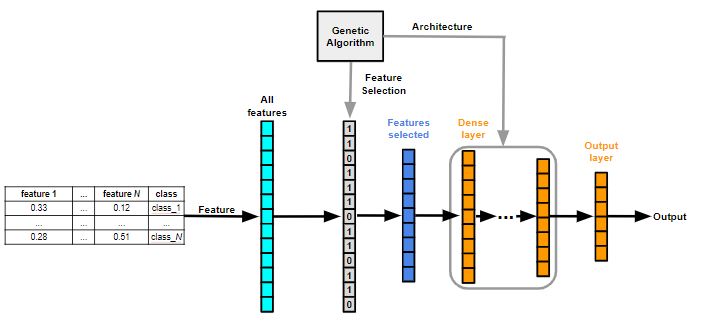
- ARCHITECTURE: MLP
- BACKBONE: GASearch
- FEATURE_SELECTION: True
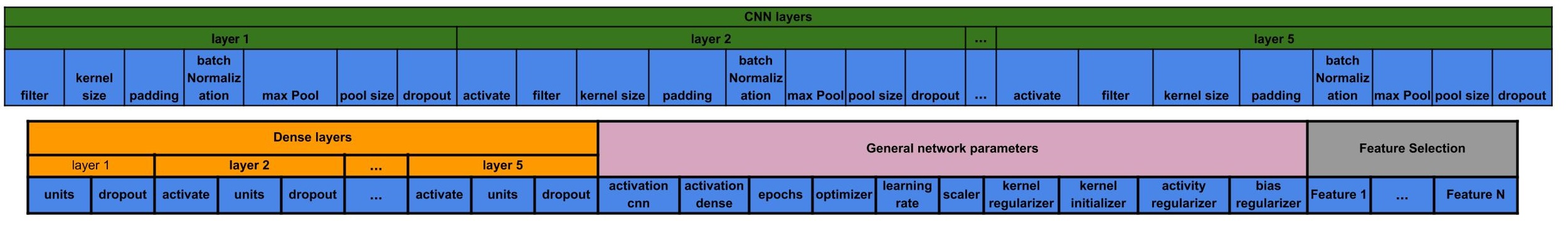
The Genetic Algorithm chromosome that encodes this model is shown below:
Parameters:
- ARCHITECTURE: MLP
- BACKBONE: GASearch
- FEATURE_SELECTION: False
The Genetic Algorithm chromosome that encodes this model is shown below:
Parameters:
- ARCHITECTURE: MLP
- BACKBONE: MLP
- FEATURE_SELECTION: True
The Genetic Algorithm chromosome that encodes this model is shown below:
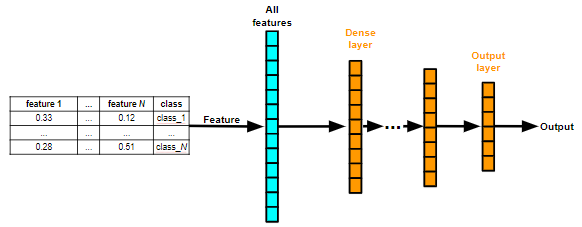
Parameters:
- ARCHITECTURE: MLP
- BACKBONE: MLP
- FEATURE_SELECTION: False
Parameters:
- ARCHITECTURE: CNN
- BACKBONE: GASearch
The Genetic Algorithm chromosome that encodes this model is shown below:
Parameters:
- ARCHITECTURE: CNN
- BACKBONE: Some pre-trained models from table for classification
Parameters:
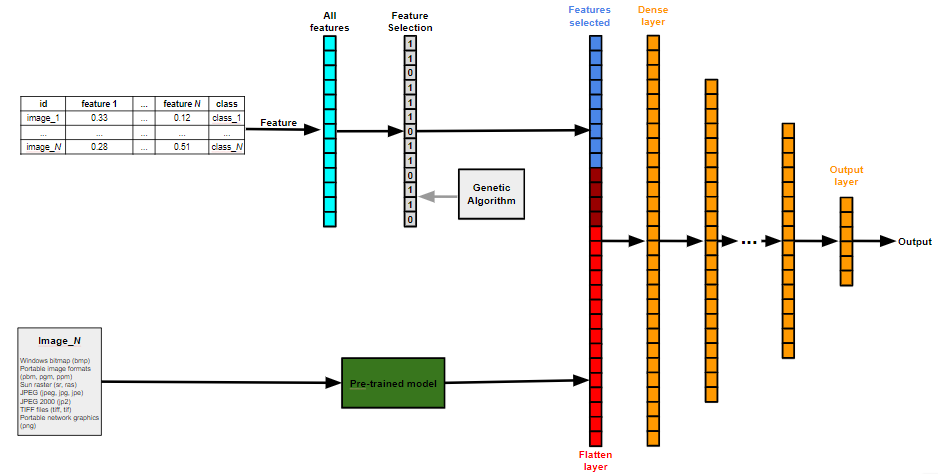
- ARCHITECTURE: CNN/MLP
- BACKBONE: GASearch
- FEATURE_SELECTION: True
The Genetic Algorithm chromosome that encodes this model is shown below:
Parameters:
- ARCHITECTURE: CNN/MLP
- BACKBONE: GASearch
- FEATURE_SELECTION: False
The Genetic Algorithm chromosome that encodes this model is shown below:
Parameters:
- ARCHITECTURE: CNN/MLP
- BACKBONE: Some pre-trained models from table for classification
- FEATURE_SELECTION: True
The Genetic Algorithm chromosome that encodes this model is shown below:
Parameters:
- ARCHITECTURE: CNN/MLP
- BACKBONE: Some pre-trained models from table for classification
- FEATURE_SELECTION: False
- TASK: Segmentation
Parameters:
- ARCHITECTURE: CNN
- BACKBONE: Some pre-trained models from table for segmentation
- FEATURE_SELECTION: False
Pay Atention: This optimization works only for classification task!!
| Parameter | activate | filter | kernel_size | padding | batch_normalization | max_pool | pool_size | dropout |
|---|---|---|---|---|---|---|---|---|
| Values | True False |
List of integers contained in the range [1:10]. The number of filters is defined by the function: filters = 2^i, where i is the provided value. | List of integers contained in the range [1:20] | valid same |
True False |
True False |
List of integers contained in the range [1:20] | List of floats contained in the range [0.0:1.0] |
| Default Values | True False |
[4, 5, 6, 7, 8, 9] | [2, 3, 4, 5, 6] | valid same |
True False |
True False |
[2, 3, 4, 5] | [0.0, 0.1, 0.2, 0.3, 0.4] |
| Description | True: this layer is added to the model False: this layer is not added to the model |
Integer, the dimensionality of the output space (i.e. the number of output filters in the convolution) | Integer, specifying the height and width of the 2D convolution window. The same value will be used for both dimensions. | Valid: means "no padding" Same: results in padding the input such that the output has the same length as the original input |
True: Normalize the activations of the previous layer at each batch, i.e. applies a transformation that maintains the mean activation close to 0 and the activation standard deviation close to 1 False: Does not do anything |
True: Applies the max pooling operation False: Does not do anything |
Integer factor by which to downscale (vertical, horizontal). The same window length will be used for both dimensions. | Dropout consists in randomly setting a fraction rate of input units to 0 at each update during training time, which helps prevent overfitting. |
| Parameter | activate | units | dropout |
|---|---|---|---|
| Values | True False |
List of integers contained in the range [1:5000] | List of floats contained in the range [0.0:1.0] |
| Default Values | True False |
[20, 50, 80, 100, 150, 200, 250, 500] | [0.0, 0.1, 0.2, 0.3, 0.4] |
| Description | True: this layer is added to the model False: this layer is not added to the model" |
The amount of neurons in the dense layer. | Dropout consists in randomly setting a fraction rate of input units to 0 at each update during training time, which helps prevent overfitting. |
| Parameter | activation_cnn | activation_dense | epochs | optimizer | learning_rate | scaler | kernel_regularizer | kernel_initializer | activity_regularizer | bias_regularizer |
|---|---|---|---|---|---|---|---|---|---|---|
| Values | List of activation functions, among the options: relu tanh elu softmax selu softplus softsign sigmoid hard_sigmoid exponential linear |
List of activation functions, among the options: relu tanh elu softmax selu softplus softsign sigmoid hard_sigmoid exponential linear |
List of integers contained in the range [1:2000] | List of optimizers, among the options: Adam SGD RMSprop Adagrad Adadelta Adamax Nadam |
List of floats contained in the range [1E-5:1.0] | List of scaler functions, among the options: Standard MinMax |
List of regularizer functions, among the options: None l1 l2 l1_l2 |
List of inicializer functions, among the options: Zeros Ones RandomNormal RandomUniform TruncatedNormal VarianceScaling Orthogonal lecun_uniform glorot_normal glorot_uniform he_normal lecun_normal he_uniform |
List of regularizer functions, among the options: None l1 l2 l1_l2 |
List of regularizer functions, among the options: None l1 l2 l1_l2 |
| Default Values | ['relu', 'tanh'] | ['relu', 'tanh'] | [30, 45, 60, 90, 120, 150, 200] | ['Adam', 'SGD'] | [0.0001, 0.001, 0.01, 0.05, 0.1] | ['Standard', 'MinMax'] | [None] | ['glorot_uniform'] | [None] | [None] |
| Description | Activation function to use in the CNN layers. | Activation function to use in the Dense layers. | Number of epochs to train the model. | Optimizer used in training the model. | Learning rate used in training the model. | Standardization method used to scale the data | Regularizer function applied to the kernel weights matrix. https://keras.io/regularizers/ |
Initializer for the kernel weights matrix. https://keras.io/initializers/ |
Regularizer function applied to the output of the layer (its activation). https://keras.io/regularizers/ |
Regularizer function applied to the bias vector. https://keras.io/regularizers/ |
Example: Classification dataset.
There is a standard way to organize your images to train a classification model. After collecting your images, you should first classify them by data set, such as training and validation, and second by classes. In each of the folder directories of the data set, we will have subdirectories, one for each class where the images will be placed.
The directory structure should look like this:
data/train/
├── class_1/
├── image0001.jpg
├── ...
├── ...
├── class_N/
├── image0001.jpg
├── ...
data/val/
├── class_1/
├── image0001.jpg
├── ...
├── ...
├── class_N/
├── image0001.jpg
├── ...
The CSV structure should look like this:
| id | feature 1 | feature 2 | ... | feature N | class |
|---|---|---|---|---|---|
| image_001 | 0.33 | 0.06 | ... | 0.12 | class_1 |
| image_002 | 0.31 | 0.38 | ... | 0.74 | class_1 |
| image_003 | 0.28 | 0.56 | ... | 0.18 | class_2 |
| image_004 | 0.02 | 0.78 | ... | 0.65 | class_2 |
| image_005 | 0.97 | 0.67 | ... | 0.13 | class_3 |
| ... | ... | ... | ... | ... | ... |
| image_N | 0.28 | 0.39 | ... | 0.51 | class_N |
The file can be sep = ',' and decimal point = '.':
id, feature 1, feature 2, ..., feature N, class
image_001, 0.33, 0.06, ..., 0.12, class_1
image_002, 0.31, 0.38, ..., 0.74, class_1
image_003, 0.28, 0.56, ..., 0.18, class_2
image_004, 0.02, 0.78, ..., 0.65, class_2
image_005, 0.97, 0.67, ..., 0.13, class_3
image_N, 0.28, 0.39, ..., 0.51, class_N Or the file can be sep = ';' and decimal point = ',':
id; feature 1; feature 2; ...; feature N; class
image_001; 0,33; 0,06; ...; 0,12; class_1
image_002; 0,31; 0,38; ...; 0,74; class_1
image_003; 0,28; 0,56; ...; 0,18; class_2
image_004; 0,02; 0,78; ...; 0,65; class_2
image_005; 0,97; 0,67; ...; 0,13; class_3
image_N; 0,28; 0,39; ...; 0,51; class_N Example: Segmentation dataset.
data/train/
├── image/
├── image0001.jpg
├── ...
├── mask/
├── image0001.jpg
├── ...
data/val/
├── image/
├── image0001.jpg
├── ...
├── mask/
├── image0001.jpg
├── ...
Each image and its respective mask should be the same name.
Please, read the getting-started.md for the basics.
MIT