MOCA has been validated with the following dependencies:
- Python (3.10)
- numpy (==1.26.3)
- scipy (==1.12.0)
- matplotlib (==3.8.0)
- summa (==1.0.0)
Assuming that Python 3.10 is installed on your system, we'll
start by setting up a virtual environment named env and activating
python3.10 -m venv env
source env/bin/activate
The prompt on the terminal should update after activation so
that it is prefaced with (env). If not a mistake has occured.
python -m pip install --upgrade pip
Now install the summa package from the pySUMMA GitHub
repository. If the dependencies numpy, scipy, and matplotlib
are not already installed, they will automatically be installed
with summa
python -m pip install git+https://github.com/robert-vogel/pySUMMA.git
Next, and lastly, install moca from the GitHub repo:
python -m pip install git+https://github.com/robert-vogel/moca.git
Here we are going to use moca simulation tools to
demonstrate both unsupervised and supervised greedy
selection moca. Assuming the dependencies are installed
import numpy as np
import matplotlib.pyplot as plt
from moca import simulate, stats
from moca import classifiers as clsThen let's simulate data
m_classifiers = 15
n_samples = 1000
n_positives = 300
auc_lims = (0.45, 0.75)
seed = 42
rng = np.random.default_rng(seed=seed)
auc = rng.uniform(low=auc_lims[0],
high=auc_lims[1],
size=m_classifiers)
corr_matrix = simulate.make_corr_matrix(m_classifiers,
independent=True,
seed=rng)
data, labels = simulate.rank_scores(n_samples,
n_positives,
auc,
corr_matrix,
seed=rng)Now with our simulation data, we can infer the optimal
weights using the Umoca class
cl = cls.Umoca()
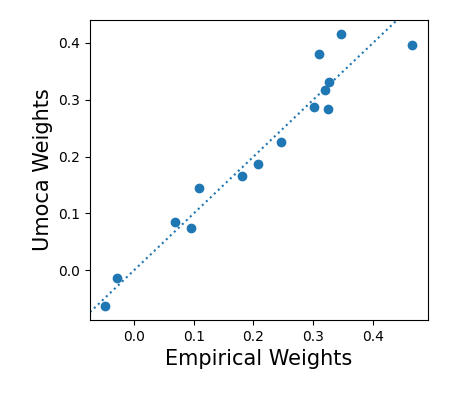
cl.fit(data)The inferred moca weights should be proportional to the
empirical signal-to-noise ratio. Using the stats module,
the signal-to-noise ratio is
true_weights = np.zeros(m_classifiers)
for i in range(m_classifiers):
true_weights[i] = stats.snr(data[i, :], labels)
true_weights /= stats.l2_vector_norm(true_weights)and plotting
fig, ax = plt.subplots(1,1, figsize=(4.5, 4))
ax.scatter(true_weights, cl.weights)
ax.axline((0,0), slope=1, linestyle=":")
ax.set_xlabel("Empirical Weights", fontsize=15)
ax.set_ylabel("Umoca Weights", fontsize=15)
ax.set_position((0.2, 0.2, 0.75, 0.75))