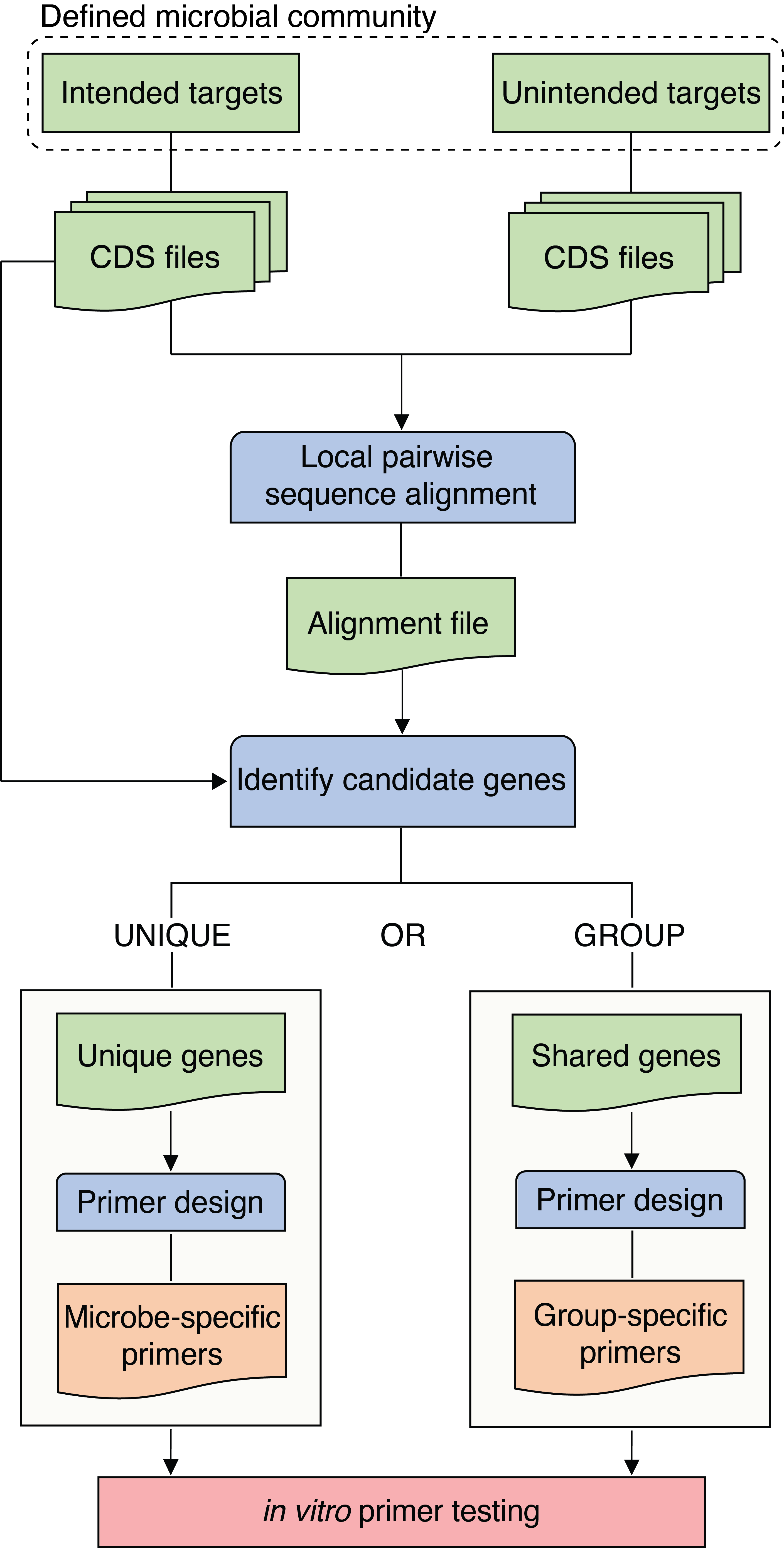
PUPpy (Phylogenetically Unique Primers in python) is a fully automated pipeline to design taxon-specific primers for any defined bacterial community.
PUPpy can design both microbe-specific primers, which selectively amplify individual members of a community, and group-specific primers, which selectively amplify user-selected members.
PUPpy-designed primers can be used to:
- Detect microbes (e.g. with PCR),
- Quantify substrain-level absolute microbial abundance (qPCR/ddPCR), and
- Any other primer uses
- Phylogenetically Unique Primers in python (PUPpy)
- Table of contents
- Installation
- Important: before you start
- How it works
- Usage
PUPpy is currently ONLY available for MacOS and Linux
Installation of the puppy package varies depending on the architecture of your computer. To check what you have, open your terminal app and run: uname -m. The architectures currently supported are:
- x86-64 (Mac Intel chips, or emulated)
- linux-64
We are currently working on making puppy also available on arm64 (Mac M1/M2 chips). For more information on how M1/M2 users can emulate the osx-64 architecture (and thus install puppy) please see the links below:
- https://stackoverflow.com/questions/71515117/how-to-set-up-a-conda-osx-64-environment-on-arm-mac
- https://taylorreiter.github.io/2022-04-05-Managing-multiple-architecture-specific-installations-of-conda-on-apple-M1/
Ensure that the conda-forge and bioconda channels are added prior to installation:
conda config --add channels defaults
conda config --add channels conda-forge
conda config --add channels biocondaYou can now create a new environment and install the puppy package:
conda create -n puppy -c hghezzi -y puppyActivate your environment prior to use:
conda activate puppyCURRENTLY IN PROGRESS...
You can set up the conda environment to run PUPpy using the YAML definition found in this repository:
# Clone PUPpy GitHub directory
git clone https://github.com/Tropini-lab/PUPpy.git
# Change directory
cd PUPpy
# Create and set up conda environment
conda deactivate
conda env create -f puppy_env.yml
conda activate PUPpy-
PUPpy was developed to design taxon-specific primers in DEFINED bacterial communities.
While in limiting cases it may be possible to use PUPpy-designed primers in undefined communities, specificity cannot be ensured in silico with PUPpy.
-
Primers should always be tested in vitro prior to use.
PCR can be misterious, and while primers may look perfect in silico, we strongly encourage confirming their specifity in vitro prior to use.
-
Read the "INPUT" section below to ensure cds files are named correclty prior to use
PUPpy takes any number of bacterial CDS files as input. Input CDS files are aligned against each other using MMseqs2 and then parsed to identify candidate unique or group-specific genes within the defined bacterial community provided by the user. Taxon-specific primers are then designed using Primer3 and provided as output in a tsv file.
IMPORTANT: If installing by cloning the GitHub directory, make sure you are in the scripts directory any time you are running PUPpy scripts or it won't work.
cd ./PUPpy/scripts
PUPpy operates in 2 main steps:
puppy-align- performs local pairwise sequence alignment of all the input genes against each other, andpuppy-primers- designs taxon-specific primers based on user-determined parameters.
Detailed usage information, including all the primer design parameters, can be seen by running -h or --help at each step.
puppy-align -h
puppy-primers -h
The alignment step must always be run first for any new defined bacterial community.
puppy-align --pr <PATH>/test/intended_input -nt <PATH>/test/unintended_input -o <PATH>/test/alignment_outputThis command creates the output file <PATH>/test/alignment_output/ResultDB.tsv which can be used as input for the primer design command (step 2). The command puppy-primers can be run as many times as desired without having to rerun puppy-align again, as long as the bacterial community remains unchanged.
The second step consists in designing taxon-specific primers unique to individual members or shared by groups in the bacterial community.
puppy-primers -pr <PATH>/test/intended_input -i <PATH>/test/alignment_output/ResultDB.tsv -o <PATH>/test/unique_output
By default, puppy-primers outputs unique primers. To design group primers, add the argument -p group to the code above.
puppy-primers requires 2 arguments as input:
-
-pror--PRIMERTARGET- the same folder aspuppy-align, containing the CDS files of the organisms for which you want to design taxon-specific primers. -
-ior--input- either the alignment file,ResultDB.tsvorUniqueGenesList.tsvUniqueGenesList.tsvis a file created by runningpuppy-primerson unique mode, containing the list of unique genes found for the organisms listed in--target_species.- This is a shortcut if you need to run
puppy-primersmultiple times on the same community and it provides the same output as usingResultDB.tsv. The only difference is that you can only useUniqueGenesList.tsvafter having runpuppy-primersat least once before, whileResultDB.tsvmust be used immediately afterpuppy-align.
You can see the default primer design parameters used by Primer3 by running puppy-primers -h.
Command line options for puppy-align
General:
--help This help
--primerTarget [X] Directory with the CDS files of the intended targets in the defined microbial community, for which primers should be designed (default '')
--nonTarget [X] Directory with CDS files of unintended targets in the defined microbial community, for specificity checks (default '')
--outdir [X] Output directory (default 'Align_OUT')
--identity [X] Identity thresholds to report sequence alignments by MMseqs2 (default '0.3')
Command line options for puppy-primers
General:
--help This help
--primers_type [X] Design unique or shared primers among the target bacterial group (default 'unique')
--primerTarget [X] Directory containing the CDS files for the species to design taxon-specific primers (default '')
--input [X] Input file to generate primers. Either 'ResultDB.tsv' OR 'UniqueGenesList.tsv' file must be provided (default '')
--outdir [X] Relative path to the output folder (default 'Primer3_output')
Primer3 parameters:
--genes_number [X] Number of genes per species to design primers (default '5')
--primers_number [X] Number of primer pairs to design for each gene (default '4')
--optimal_primer_size [X] Primer optimal size (default '20')
--min_primer_size [X] Primer minimum size (default '18')
--max_primer_size [X] Primer maximum size (default '22')
--optimal_primer_Tm [X] Primer optimal melting temperature (default '60.0')
--min_primer_Tm [X] Primer minimum melting temperature (default '58.0')
--max_primer_Tm [X] Primer maximum melting temperature (default '63.0')
--max_Tm_diff [X] Maximum Tm difference between the primer pair (default '2.0')
--min_primer_gc [X] Primer minimum GC content (default '40.0')
--max_primer_gc [X] Primer maximum GC content (default '60.0')
--product_size_range [X] Product size range (default '75 150')
--max_poly_x [X] Maximum poly X allowed (default '3')
--GC_clamp [X] Primer GC clamp (default '1')
Currently, PUPpy supports CDS files generated from any of these 3 approaches: prokka, RAST and/or downloaded from the NCBI. This is necessary because PUPpy only recognises FASTA headers formats from these 3 programs.
- For Prokka, rename the
.ffnoutput file to end with the extension.fna
Examples of accepted FASTA headers are shown here:
# CDS file downloaded from the NCBI:
>lcl|NC_004663.1_cds_WP_011107050.1_1 [locus_tag=BT_RS00005] [db_xref=GeneID:1075082] [protein=hypothetical protein] [protein_id=WP_011107050.1] [location=93..710] [gbkey=CDS]
# CDS file from prokka:
>COAIMFFE_00001 putative protein
# CDS file from RAST:
>fig|6666666.855680.peg.1
Moreover, input CDS filenames must meet the following 3 requirements to be used by PUPpy:
-
Filename must start with a unique identifier that allows you to distinguish organisms in the defined community.
- e.g.
Bacteroides_theta_VPI5482
- e.g.
-
Filename must contain the string cds.
- e.g.
cds,cds_from_genomic,cds_genomicetc...
- e.g.
-
Filename must end with the extension
.fna- e.g.
cds.fna,cds_from_genomic.fna,cds_genomic.fna, etc...
- e.g.
Examples of accepted CDS filenames:
B_theta_VPI5482_cds.fnaBacteroides_thetaiotaomicron_VPI_5482_cds_from_genomic.fna
The key output of puppy-align is the file ResultDB.tsv, which stores exhaustive information about all the local pairwise alignments. To see an example of this output file, go to: ./test/alignment_output/ResultDB.tsv in this repository.
The outputs of puppy-primers vary depending on which mode is run:
Unique mode:
Stats_pipelineOutput.tsv- table containing the number of unique genes found and total number of genes for each community member.UniqueGenesPlot.pdf- barplot showing the number of unique genes found for each community member.UniqueGenesList.tsv- list of unique genes found for each memberUniquePrimerTable.tsv- output table with the taxon-specific primers designed and their respective parametersprimer3_files/- folder containing the individual primer3 outputs of the primers inUniquePrimerTable.tsv
Examples of these outputs can be seen in this repository at ./test/unique_output/
Group mode:
-
GroupPrimerTable.tsv- output table with the taxon-specific primers designed and their respective parameters -
primer3_files/- folder containing the individual primer3 outputs of the primers inGroupPrimerTable.tsv -
IdealGroupGenes.tsv- List of most ideal candidate genes used by PUPpy to design group-specific primers. Ideal genes must meet the following requirements:- The candidate gene has exactly 1 alignment to each intended target;
- The candidate gene only amplifies intended species in the defined community;
- The candidate gene aligns perfectly (100% ID) to each target gene;
- The entire length of the candidate gene (i.e. 100% query coverage) aligns to each target gene;
- The candidate gene aligns to the entire sequence of each target gene (i.e. 100% target coverage).
-
SecondChoiceGroupGenes.tsv- List of not-ideal genes that will be used by PUPpy to design group-specific primers only if no ideal genes are found. "Second choice" genes must meet the following requirements:- The candidate gene has more than 1 alignment to at least one intended target;
- The candidate gene only amplifies intended species in the defined community;
- The candidate gene does not align perfectly to at least one target gene;
- Only a portion of the candidate gene (i.e. <100% query coverage) aligns to at least one target gene;
- The candidate gene aligns does not align to the entire sequence of at least one target gene.
-
UndesiredGroupGenes.tsv- List of genes that will not be considered by PUPpy, as they would not yield group-specific primers. Undesired genes must meet the following requirements:- The candidate gene has more than 1 alignment to at least one intended target and it does not amplify all targets OR it does not amplify any intended targets;
- The candidate gene amplifies unintended species in the defined community;
- The candidate gene not align perfectly to at least one target gene;
- Only a portion of the candidate gene (i.e. <100% query coverage) aligns to at least one target gene;
- The candidate gene aligns does not align to the entire sequence of at least one target gene.
Examples of these outputs can be seen in this repository at ./test/group_output/
- Hans Ghezzi
- Katharine Michelle Ng
- Juan Camilo Burckhardt
- Yiyun Michelle Fan
If you use PUPpy in your research, please cite the original paper: https://www.biorxiv.org/content/10.1101/2023.12.18.572184v1
PUPpy is made available under GPLv3. See LICENSE for details. Copyright Carolina Tropini.
Developed by Hans Ghezzi at the University of British Columbia (UBC).