a library to compute intersections between mesh cells and implicit (level set) functions using marching cubes/tetrahedra algorithms.
The current version supports intervals, triangles and tetrahedra. Future versions will include quadrilaterals, hexahedra, pyramids and prisms.
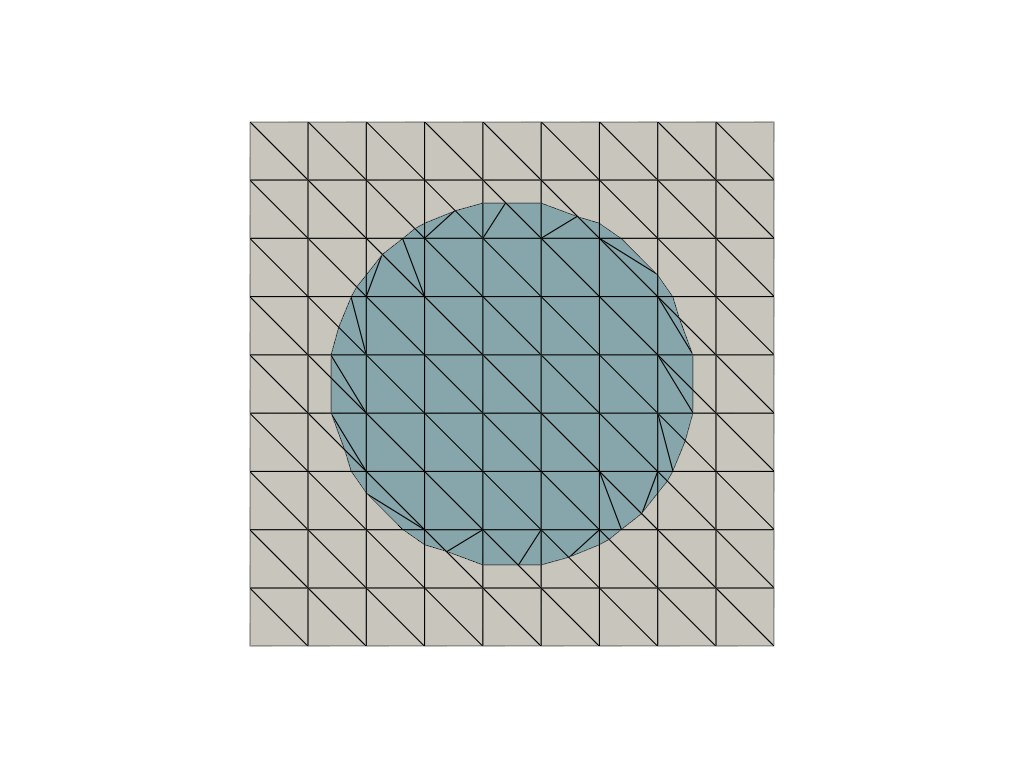
The library contains utility functions to cut through vtk meshes using pyvista. It is also used as a basis to generate run time quadrature rules in immersed boundary methods/cut finite element methods (see CutFEMx).
The images below show a cut through a vtk mesh using pyvista and CutCells.
The corresponding demonstration code is located in python/demo/cut_vtk_mesh.
To install the CutCells library, first install the C++ library and then build the python interface
In the cpp/ directory:
cmake -DCMAKE_BUILD_TYPE=Release -B build-dir -S .
cmake --build build-dir
cmake --install build-dirYou may need to use sudo for the final install step. Using the CMake
build type Release is recommended for performance.
After installing the C++ library, install the Python interface by running in
the directory python/:
cmake -DCMAKE_BUILD_TYPE=Release -B build-dir -S .
cmake --build build-dir
cmake --install build-dirand then
python3 -m pip install .There are demons for both the C++ and the python interface.
For the C++ interface the demos are located in cpp/demo. The C++ demos are built with
cmake -DCMAKE_BUILD_TYPE=Release -B build-dir -S .
cmake --build build-dir
cmake --install build-dirin the corresponding demo folder, e.g. cpp/demo/cut_triangle
The python demos are located in python/demo.
CutCells requires a C++20 compiler and depends on the C++ standard template library.
For the python interface, CutCells requires nanobind.
At runtime for the python examples, CutCells requires numpy and pyvista for visualizations.
The library contains python pytest tests in python/tests.