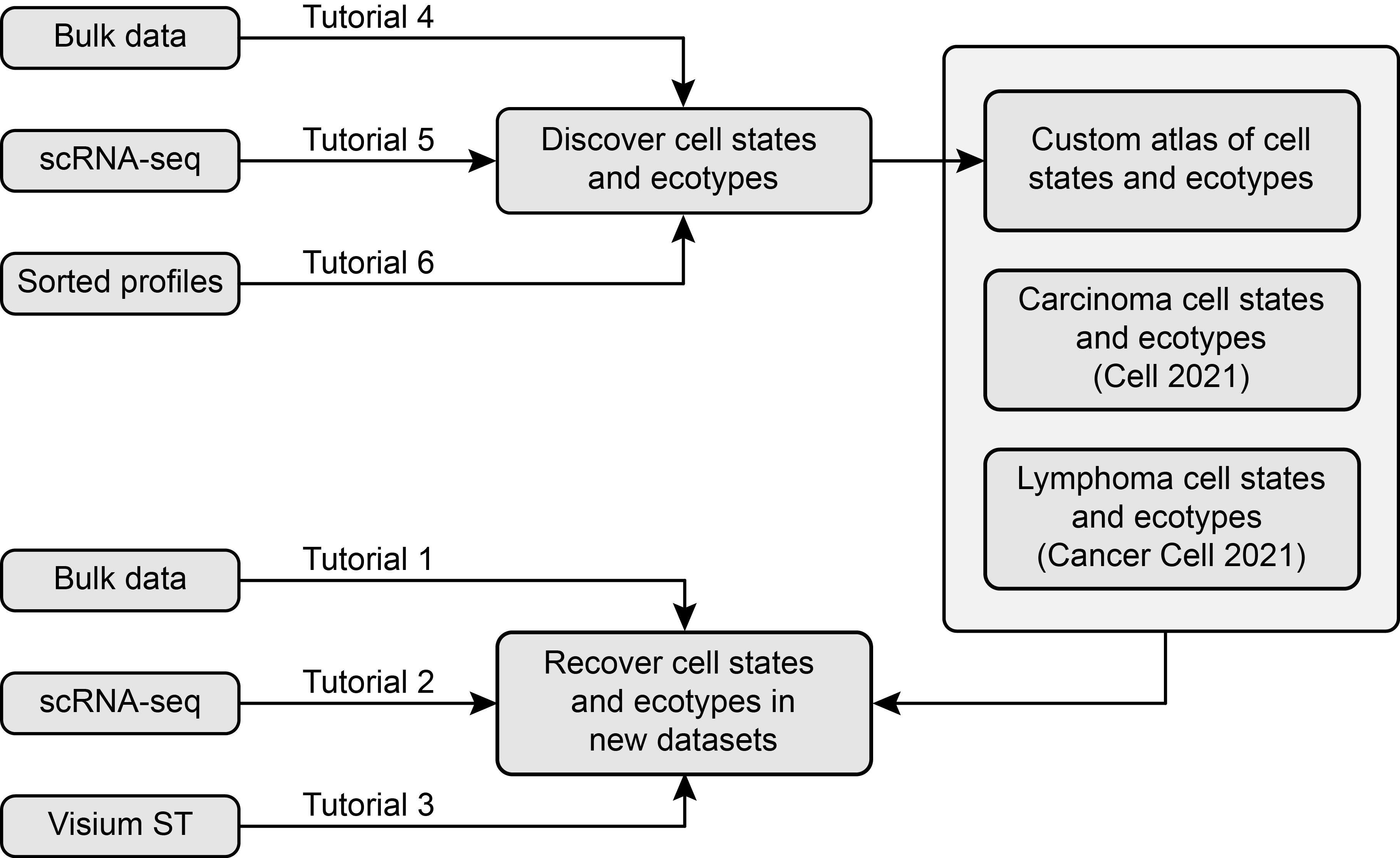
EcoTyper is a machine learning framework for large-scale identification of cell type-specific transcriptional states and their co-association patterns from bulk and single-cell (scRNA-seq) expression data.
We have already defined cell states and ecotypes across carcinomas (Luca/Steen et al., Cell 2021) and in diffuse large B cell lymphoma (DLBCL) (Steen/Luca et al., Cancer Cell 2021). The current version of EcoTyper allows users to recover the cell states and ecotypes for these two tumor categories in their own data. Additionally, it allows users to discover and recover cell states and ecotypes in their system of interest, including directly from scRNA-seq data (see Tutorial 5). Below we illustrate each of these functionalities.
If EcoTyper software, data, and/or website are used in your publication, please cite the following paper(s):
- Luca/Steen et al., Cell 2021 (detailed description of EcoTyper and application to carcinomas).
- Steen/Luca et al., Cancer Cell 2021 (application of EcoTyper to lymphoma).
The latest version of EcoTyper source code can be found on EcoTyper GitHub repository and Ecotyper website. To set up EcoTyper, please download this folder locally:
git clone https://github.com/digitalcytometry/ecotyper
cd ecotyperor:
wget https://github.com/digitalcytometry/ecotyper/archive/refs/heads/master.zip
unzip master.zip
cd ecotyper-masterThe R packages listed below are required for running EcoTyper. The version numbers indicate the package versions used for developing and testing the EcoTyper code. Other R versions might work too:
- R (v3.6.0 and v4.1.0).
- R packages: ComplexHeatmap (v2.2.0 and v2.8.0), NMF (v0.21.0 and v0.23.0), RColorBrewer (v1.1.2), cluster (v2.1.0 and v2.1.2)), circlize (v0.4.10 and v0.4.12), cowplot (v1.1.0 and v1.1.1), data.table (base package R v3.6.0 and v4.1.0), doParallel (v1.0.15 and v1.0.16), ggplot2 (v3.3.2, v3.3.3), grid (base package R v3.6.0 and v4.1.0), reshape2 (v1.4.4), viridis (v0.5.1 and v0.6.1), config (v0.3.1), argparse (v2.0.3), colorspace (v1.4.1 and v2.0.1), plyr (v1.8.6).
These packages, together with the other resources pre-stored in the EcoTyper folder, allow users to:
- perform the recovery of previously defined cell states and ecotypes in their own bulk RNA-seq, microarray and scRNA-seq data (Tutorials 1 and 2).
- perform cell state and ecotype discovery in scRNA-seq and pre-sorted cell type-specific profiles (Tutorials 5 and 6).
Besides these packages, the additional resources described in the next section are needed for analyses described in Tutorials 3 and 4.
For some use cases, such as cell state and ecotype recovery in spatial transcriptomics assays (Tutorial 3) and de novo identification of cell states and ecotypes from bulk expression data (Tutorial 4), EcoTyper relies on CIBERSORTx (Newman et al., Nature Biotechnology 2019, a digital cytometry framework for enumerating cell types in bulk data and performing in silico deconvolution of cell type specific expression profiles. In these situations, the following additional resources are needed for running EcoTyper:
- Docker.
- CIBERSORTx executables than can be downloaded from the CIBERSORTx website, as Docker images. Specifically, EcoTyper requires the CIBERSORTx Fractions and CIBERSORTx HiRes modules. Please follow the instructions on the Download section of the website to download the Docker images and obtain the Docker tokens necessary for running them.
EcoTyper is a standalone software, implemented in R (not an R package). Some of the EcoTyper functions are computationally intensive, especially for the cell state discovery step described in Tutorials 4-6. Therefore, EcoTyper is designed as a collection of modular command-line R scripts, that can be run in parallel on a multi-processor server or a high-performance cluster. Each script is designed such that its instances can typically be run on a single core.
We provide wrappers over these scripts that encapsulate the typical EcoTyper workflows (Tutorials 1-6). These wrappers can be run on a multi-core system, and allow users to discover cell states and ecotypes in their own bulk, scRNA-seq and FACS-sorted data, as well as recover previously discovered cell states and ecotypes in bulk tissue expression profiles, spatial transcriptomics assays, and single-cell RNA-seq data.
EcoTyper performs two major types of analysis: discovery of cell states and ecotypes, starting from bulk, scRNA-seq and pre-sorted cell type specific expression profiles (e.g. FACS-sorted or deconvolved in silico); and recovery of previously defined cell states and ecotypes in new bulk, scRNA-seq and spatial transcriptomics data.
When the input is bulk data, EcoTyper performs the following major steps for discovering cell states and ecotypes:
- In silico purification: This step enables imputation of cell type-specific gene expression profiles from bulk tissue transcriptomes, using CIBERSORTx (Newman et al., Nature Biotechnology 2019).
- Cell state discovery: This step enables identification and quantitation of cell type-specific transcriptional states.
- Ecotype discovery: This step enables co-assignment of cell states into multicellular communities (ecotypes).
When the input is scRNA-seq or bulk-sorted cell type-specific profiles (e.g., FACS-purified), EcoTyper performs the following major steps for discovering cell states and ecotypes:
- Gene filtering: This step filters out genes that do not show cell type specificity.
- Cell state discovery: This step enables identification and quantitation of cell type-specific transcriptional states.
- Ecotype discovery: This step enables co-assignment of cell states into multicellular communities (ecotypes).
Regardless of the input type used for deriving cell states and ecotypes, EcoTyper can perform cell state and ecotype recovery in external expression datasets. The recovery can be performed in bulk, scRNA-seq and spatial transcriptomics data.
We provide 6 tutorials illustrating these functionalities. The first three demonstrate how the recovery of cell states and ecotypes can be performed with various input types. The last three demonstrate how the recovery of cell states and ecotypes can be performed with various input types:
- Tutorial 1: Recovery of Cell States and Ecotypes in User-Provided Bulk Data
- Tutorial 2: Recovery of Cell States and Ecotypes in User-Provided scRNA-seq Data
- Tutorial 3: Recovery of Cell States and Ecotypes in Visium Spatial Gene Expression Data
- Tutorial 4: De novo Discovery of Cell States and Ecotypes in Bulk Expression Data
- Tutorial 5: De novo Discovery of Cell States and Ecotypes in scRNA-seq Data
- Tutorial 6. De novo Discovery of Cell States and Ecotypes in Pre-Sorted Data
A schema of the tutorials is presented below:
EcoTyper comes pre-loaded with the resources necessary for the reference-guided recovery of cell states and ecotypes previously defined in carcinoma or lymphoma, in user-provided bulk expression data. In the carcinoma EcoTyper paper, we demonstrate that prior deconvolution of bulk data using CIBERSORTx HiRes is not necessary for high-fidelity recovery of cell states in bulk-tissue expression data. We can proceed to the recovery of states based on bulk data only.
In this tutorial, we illustrate how EcoTyper can be used to recover the cell states and ecotypes that we defined across carcinomas and in diffuse large B cell lymphoma (DLBCL), in a set of the bulk samples from lung adenocarcinoma (LUAD) from TCGA and bulk samples from diffuse large-cell lymphoma (DLBCL), respectively. Plese note that the recovery procedure described in this tutorial can also be applied on user-defined cell states and ecotypes, derived as described in Tutorials 4-6.
For this section, we used a subset of the TCGA bulk samples from lung
adenocarcinoma (LUAD), available in example_data/bulk_lung_data.txt,
together with the sample annotation file
example_data/bulk_lung_annotation.txt.
The script used to perform recovery in bulk data is called
EcoTyper_recovery_bulk.R:
Rscript EcoTyper_recovery_bulk.R -h## usage: EcoTyper_recovery_bulk.R [-d <character>] [-m <PATH>] [-a <PATH>]
## [-c <character>] [-t <integer>] [-o <PATH>]
## [-h]
##
## Arguments:
## -d <character>, --discovery <character>
## The name of the discovery dataset used to define cell
## states and ecotypes. Accepted values: 'Carcinoma' will
## recover the cell states and ecotypes defined across
## carcinomas, as described in the EcoTyper carcinoma
## paper, 'Lymphoma' will recover the cell states and
## ecotypes defined in diffuse large B cell lymphoma
## (DLBCL), as described in the EcoTyper lymphoma paper,
## '<MyDiscovery>' the value used in the field 'Discovery
## dataset name' of the config file used for running
## EcoTyper discovery ('EcoTyper_discovery.R') script.
## [default: 'Carcinoma']
## -m <PATH>, --matrix <PATH>
## Path to a tab-delimited file containing the input bulk
## tissue expression matrix, with gene names on the first
## column and sample ids as column names [required].
## -a <PATH>, --annotation <PATH>
## Path to a tab-delimited annotation file containing the
## annotation of samples in the input matrix. This file
## has to contain in column 'ID' the same ids used as
## column names in the input matrix, and any number of
## additional columns. The additional columns can be
## plotted as color bars in the output heatmaps.
## [default: 'NULL']
## -c <character>, --columns <character>
## A comma-spearated list of column names from the
## annotation file to be plotted as color bar in the
## output heatmaps. [default: 'NULL']
## -t <integer>, --threads <integer>
## Number of threads. [default: '10']
## -o <PATH>, --output <PATH>
## Output directory path. [default: 'RecoveryOutput']
## -h, --help Print help message.
The script takes the following arguments:
-
-d/–discovery: The name of the discovery dataset used for defining cell states. By default, the only accepted values are Carcinoma and Lymphoma (case sensitive), which will recover the cell states that we already defined across carcinomas and in lymphoma, respectively. If the user defined cell states in their own data (Tutorials 4-6), the name of the discovery dataset is the value provided in the Discovery dataset name field of the configuration file used for running cell state discovery. In our tutorial, the name of the discovery dataset is Carcinoma.
-
-m/–matrix: Path to the input expression matrix. The expression matrix should be in the TPM or FPKM space for bulk RNA-seq and non-logarithmic (exponential) space for microarrays. It should have gene symbols on the first column and gene counts for each sample on the next columns. Column (sample) names should be unique. Also, we recommend that the column names do not contain special characters that are modified by the R function make.names, e.g. having digits at the beginning of the name or containing characters such as space, tab or -. The lung cancer scRNA-seq data used in this tutorial looks as follows:
data = read.delim("example_data/bulk_lung_data.txt", nrow = 5)
head(data[,1:5])## Gene TCGA.37.A5EN.01A.21R.A26W.07 TCGA.37.4133.01A.01R.1100.07
## 1 A1BG 18.6400165 18.196602709
## 2 A1CF 0.0338368 0.002095014
## 3 A2M 54.1463351 35.714991125
## 4 A2ML1 4.9953315 2.383752067
## 5 A3GALT2 0.0438606 0.000000000
## TCGA.77.7465.01A.11R.2045.07 TCGA.34.5240.01A.01R.1443.07
## 1 24.83635354 23.579201761
## 2 0.02301987 0.004186634
## 3 80.63633736 86.804257397
## 4 4.08688641 3.015307103
## 5 0.00000000 0.000000000
- -a/–annotation: Path to a tab-delimited annotation file (not required). If provided, this file should contain a column called ID with the same values as the columns of the expression matrix. Additionally, this file can contain any number of columns, that can be used for plotting as color bars in the output heatmaps (see argument -c/–columns).
data = read.delim("example_data/bulk_lung_annotation.txt")
head(data)## ID Tissue Histology Type OS_Time
## 1 TCGA.37.A5EN.01A.21R.A26W.07 Tumor LUSC Primary Solid Tumor 660
## 2 TCGA.37.4133.01A.01R.1100.07 Tumor LUSC Primary Solid Tumor 238
## 3 TCGA.77.7465.01A.11R.2045.07 Tumor LUSC Primary Solid Tumor 990
## 4 TCGA.34.5240.01A.01R.1443.07 Tumor LUSC Primary Solid Tumor 1541
## 5 TCGA.05.4249.01A.01R.1107.07 Tumor LUAD Primary Solid Tumor 1523
## 6 TCGA.62.8398.01A.11R.2326.07 Tumor LUAD Primary Solid Tumor 444
## OS_Status
## 1 0
## 2 0
## 3 0
## 4 0
## 5 0
## 6 1
-
-c/–columns: A comma-separated list of column names from the annotation file (see argument -a/–annotation) to be plotted as color bars in the output heatmaps. By default, the output heatmaps contain as color bar the cell state label each cell is assigned to. The column names indicated by this argument will be added to that color bar.
-
-t/–threads: Number of threads. Default: 10.
-
-o/–output: Output folder. The output folder will be created if it does not exist.
The command line for recovering the carcinoma cell states and ecotypes in the example bulk data is:
Rscript EcoTyper_recovery_bulk.R -d Carcinoma -m example_data/bulk_lung_data.txt -a example_data/bulk_lung_annotation.txt -c Tissue -o RecoveryOutputThe output of this script for each cell type includes:
- The abundance (fraction) of each cell state in each sample:
data = read.delim("RecoveryOutput/bulk_lung_data/Fibroblasts/state_abundances.txt")
head(data[,1:5])## TCGA.37.A5EN.01A.21R.A26W.07 TCGA.37.4133.01A.01R.1100.07
## S01 2.083264e-15 0.018526046
## S02 1.379863e-01 0.012301783
## S03 1.296628e-01 0.003181937
## S04 7.119142e-03 0.003346075
## TCGA.77.7465.01A.11R.2045.07 TCGA.34.5240.01A.01R.1443.07
## S01 0.05884281 0.01150542
## S02 0.18379510 0.05917894
## S03 0.07466737 0.06311084
## S04 0.05295505 0.03820861
## TCGA.05.4249.01A.01R.1107.07
## S01 0.35434105
## S02 0.28080083
## S03 0.11240927
## S04 0.04851547
- The assignment of samples to the state with highest abundance. If the cell state with the highest abundance is one of the cell states filtered by the automatic QC filters of EcoTyper, the sample is considered unassigned and filtered out from this table. For more information about the sample filtering procedure please see the Cell state quality control section of the EcoTyper paper methods:
data = read.delim("RecoveryOutput/bulk_lung_data/Fibroblasts/state_assignment.txt")
head(data[,c("ID", "State")])## ID State
## 1 TCGA.05.4249.01A.01R.1107.07 S01
## 2 TCGA.50.6590.01A.12R.1858.07 S01
## 3 TCGA.55.6983.11A.01R.1949.07 S01
## 4 TCGA.69.7761.01A.11R.2170.07 S01
## 5 TCGA.93.7347.01A.11R.2187.07 S01
## 6 TCGA.73.4662.01A.01R.1206.07 S01
- Two heatmaps: the heatmap representing the expression of “marker” genes for each state (See Tutorial 4 for more details) in the discovery dataset and in the user-provided bulk dataset:
knitr::include_graphics("RecoveryOutput/bulk_lung_data/Fibroblasts/state_assignment_heatmap.png")The output for ecotypes includes:
- The abundance (fraction) of each ecotype in each sample:
assign = read.delim("RecoveryOutput/bulk_lung_data/Ecotypes/ecotype_abundance.txt")
dim(assign)## [1] 9 250
head(assign[,1:5])## TCGA.37.A5EN.01A.21R.A26W.07 TCGA.37.4133.01A.01R.1100.07
## LE1 0.16417228 0.02365489
## LE2 0.08192505 0.05046453
## LE3 0.12463032 0.61936802
## LE4 0.01515642 0.04878655
## LE5 0.16935221 0.04596270
## LE6 0.18055183 0.04292269
## TCGA.77.7465.01A.11R.2045.07 TCGA.34.5240.01A.01R.1443.07
## LE1 0.13700230 0.05801216
## LE2 0.03932840 0.04340269
## LE3 0.25247584 0.49820232
## LE4 0.10528174 0.02365418
## LE5 0.06864643 0.03466751
## LE6 0.09809911 0.05476600
## TCGA.05.4249.01A.01R.1107.07
## LE1 0.10219731
## LE2 0.05493172
## LE3 0.10202715
## LE4 0.12493246
## LE5 0.09788877
## LE6 0.10122469
- The assignment of samples to the carcinoma ecotype with the highest abundance. If the cell state fractions from the dominant ecotype are not significantly higher than the other cell state fractions in a given sample, the sample is considered unassigned and filtered out from this table. For more information about the sample filtering procedure please see the Ecotype discovery section of the EcoTyper paper methods:
discrete_assignments = read.delim("RecoveryOutput/bulk_lung_data/Ecotypes/ecotype_abundance.txt")
dim(discrete_assignments)## [1] 9 250
head(discrete_assignments[,1:5])## TCGA.37.A5EN.01A.21R.A26W.07 TCGA.37.4133.01A.01R.1100.07
## LE1 0.16417228 0.02365489
## LE2 0.08192505 0.05046453
## LE3 0.12463032 0.61936802
## LE4 0.01515642 0.04878655
## LE5 0.16935221 0.04596270
## LE6 0.18055183 0.04292269
## TCGA.77.7465.01A.11R.2045.07 TCGA.34.5240.01A.01R.1443.07
## LE1 0.13700230 0.05801216
## LE2 0.03932840 0.04340269
## LE3 0.25247584 0.49820232
## LE4 0.10528174 0.02365418
## LE5 0.06864643 0.03466751
## LE6 0.09809911 0.05476600
## TCGA.05.4249.01A.01R.1107.07
## LE1 0.10219731
## LE2 0.05493172
## LE3 0.10202715
## LE4 0.12493246
## LE5 0.09788877
## LE6 0.10122469
- A heatmap of cell state abundances across the samples assigned to ecotypes. Rows correspond to the cell states forming ecotypes, while columns correspond to the samples assigned to ecotypes:
knitr::include_graphics("RecoveryOutput/bulk_lung_data/Ecotypes/heatmap_assigned_samples_viridis.png")For this section, we used a subset of the bulk
samples from diffuse
large-cell lymphoma (DLBCL), available in
example_data/bulk_lymphoma_data.txt, together with the sample
annotation file example_data/bulk_lymphoma_annotation.txt.
The script used to perform recovery in bulk data is called
EcoTyper_recovery_bulk.R:
Rscript EcoTyper_recovery_bulk.R -h## usage: EcoTyper_recovery_bulk.R [-d <character>] [-m <PATH>] [-a <PATH>]
## [-c <character>] [-t <integer>] [-o <PATH>]
## [-h]
##
## Arguments:
## -d <character>, --discovery <character>
## The name of the discovery dataset used to define cell
## states and ecotypes. Accepted values: 'Carcinoma' will
## recover the cell states and ecotypes defined across
## carcinomas, as described in the EcoTyper carcinoma
## paper, 'Lymphoma' will recover the cell states and
## ecotypes defined in diffuse large B cell lymphoma
## (DLBCL), as described in the EcoTyper lymphoma paper,
## '<MyDiscovery>' the value used in the field 'Discovery
## dataset name' of the config file used for running
## EcoTyper discovery ('EcoTyper_discovery.R') script.
## [default: 'Carcinoma']
## -m <PATH>, --matrix <PATH>
## Path to a tab-delimited file containing the input bulk
## tissue expression matrix, with gene names on the first
## column and sample ids as column names [required].
## -a <PATH>, --annotation <PATH>
## Path to a tab-delimited annotation file containing the
## annotation of samples in the input matrix. This file
## has to contain in column 'ID' the same ids used as
## column names in the input matrix, and any number of
## additional columns. The additional columns can be
## plotted as color bars in the output heatmaps.
## [default: 'NULL']
## -c <character>, --columns <character>
## A comma-spearated list of column names from the
## annotation file to be plotted as color bar in the
## output heatmaps. [default: 'NULL']
## -t <integer>, --threads <integer>
## Number of threads. [default: '10']
## -o <PATH>, --output <PATH>
## Output directory path. [default: 'RecoveryOutput']
## -h, --help Print help message.
The script takes the following arguments:
-
-d/–discovery: The name of the discovery dataset used for defining cell states. By default, the only accepted values are Carcinoma and Lymphoma (case sensitive), which will recover the cell states that we already defined across carcinomas and in lymphoma, respectively. If the user defined cell states in their own data (Tutorials 4-6), the name of the discovery dataset is the value provided in the Discovery dataset name field of the configuration file used for running cell state discovery.
-
-m/–matrix: Path to the input expression matrix. The expression matrix should be in the TPM or FPKM space for bulk RNA-seq and non-logarithmic (exponential) space for microarrays. It should have gene symbols on the first column and gene counts for each sample on the next columns. Column (sample) names should be unique. Also, we recommend that the column names do not contain special characters that are modified by the R function make.names, e.g. having digits at the beginning of the name or containing characters such as space, tab or -. The bulk data used in this tutorial looks as follows:
data = read.delim("example_data/bulk_lymphoma_data.txt", nrow = 5)
head(data[,1:5])## GENES MS2010072001 MS2010072003 MS2010072004 MS2010072017
## 1 A1BG 319.59498 273.512399 263.81912 432.18048
## 2 A1BG_AS1 19.68925 100.372538 90.50134 19.58759
## 3 A1CF 49.99656 6.447184 51.09232 36.02929
## 4 A2M 3578.38986 3463.803236 2754.17141 1080.29716
## 5 A2M_AS1 2976.90082 102.167762 1044.81788 38.24889
- -a/–annotation: Path to a tab-delimited annotation file (not required). If provided, this file should contain a column called ID with the same values as the columns of the expression matrix. Additionally, this file can contain any number of columns, that can be used for plotting as color bars in the output heatmaps (see argument -c/–columns).
data = read.delim("example_data/bulk_lymphoma_annotation.txt")
head(data)## ID COO
## 1 MS2010072838 Unclassified
## 2 LONGS_p_DLBCL_AffyExpr_01_HG_U133_Plus_2_B03_830732 GCB
## 3 LONGS_p_DLBCL_AffyExpr_01_HG_U133_Plus_2_C11_830764 ABC
## 4 MS2010072042 ABC
## 5 MS2010072816 ABC
## 6 MS2010072921 ABC
## schmitz_labels
## 1 N1
## 2 EZB
## 3 Other
## 4 BN2
## 5 BN2
## 6 Other
-
-c/–columns: A comma-separated list of column names from the annotation file (see argument -a/–annotation) to be plotted as color bars in the output heatmaps. By default, the output heatmaps contain as color bar the cell state label each cell is assigned to. The column names indicated by this argument will be added to that color bar.
-
-t/–threads: Number of threads. Default: 10.
-
-o/–output: Output folder. The output folder will be created if doesn’t exist.
The command line for recovering the lymphoma cell states and ecotypes in the example bulk data is:
Rscript EcoTyper_recovery_bulk.R -d Lymphoma -m example_data/bulk_lymphoma_data.txt -a example_data/bulk_lymphoma_annotation.txt -c schmitz_labels,COO -o RecoveryOutputThe output of this script for each cell type includes:
- The abundance (fraction) of each cell state in each sample:
data = read.delim("RecoveryOutput/bulk_lymphoma_data/B.cells/state_abundances.txt")
head(data[,1:5])## MS2010072001 MS2010072003 MS2010072004 MS2010072017 MS2010072019
## S01 2.373248e-03 3.595618e-01 3.806641e-05 3.697861e-01 5.385940e-01
## S02 4.782425e-01 5.127354e-02 4.068437e-01 8.661441e-16 1.260096e-15
## S03 5.000272e-01 2.657648e-01 3.208192e-01 3.871464e-02 1.262404e-01
## S04 1.409287e-15 2.861731e-01 1.020793e-01 2.570570e-07 6.271926e-03
## S05 1.935706e-02 7.028482e-05 1.046191e-01 8.195354e-02 1.693508e-05
- The assignment of samples to the state with highest abundance. If the cell state with the highest abundance is one of the cell states filtered by the automatic QC filters of EcoTyper, the sample is considered unassigned and filtered out from this table. For more information about the sample filtering procedure please see the Cell state quality control section of the [EcoTyper paper] (https://doi.org/10.1016/j.cell.2021.09.014) methods:
data = read.delim("RecoveryOutput/bulk_lymphoma_data/B.cells/state_assignment.txt")
head(data[,c("ID", "State")])## ID State
## 1 MS2010072003 S01
## 2 MS2010072019 S01
## 3 MS2010072024 S01
## 4 MS2010072030 S01
## 5 MS2010072037 S01
## 6 MS2010072040 S01
- Two heatmaps: the heatmap representing the expression of “marker” genes for each state (See Tutorial 3 for more details) in the discovery dataset and in the user-provided bulk dataset:
knitr::include_graphics("RecoveryOutput/bulk_lymphoma_data/B.cells/state_assignment_heatmap.png")The output for ecotypes includes:
- The abundance (fraction) of each ecotype in each sample:
assign = read.delim("RecoveryOutput/bulk_lymphoma_data/Ecotypes/ecotype_abundance.txt")
dim(assign)## [1] 9 75
head(assign[,1:5])## MS2010072001 MS2010072003 MS2010072004 MS2010072017 MS2010072019
## LE1 0.04957052 0.08169883 0.08724611 7.549536e-02 0.039440174
## LE2 0.01006307 0.12841183 0.09457992 2.747772e-02 0.002943328
## LE3 0.05278894 0.03158038 0.07019048 2.622030e-01 0.246744970
## LE4 0.30161978 0.03494590 0.08382893 4.458162e-15 0.009809012
## LE5 0.17506202 0.19692135 0.14271831 1.226647e-01 0.137689497
## LE6 0.09394966 0.09523241 0.13293934 2.158494e-01 0.155547222
- The assignment of samples to the lymphoma ecotype with the highest abundance. If the cell state fractions from the dominant ecotype are not significantly higher than the other cell state fractions in a given sample, the sample is considered unassigned and filtered out from this table. For more information about the sample filtering procedure please see the Ecotype discovery section of the EcoTyper paper methods:
discrete_assignments = read.delim("RecoveryOutput/bulk_lymphoma_data/Ecotypes/ecotype_abundance.txt")
dim(discrete_assignments)## [1] 9 75
head(discrete_assignments[,1:5])## MS2010072001 MS2010072003 MS2010072004 MS2010072017 MS2010072019
## LE1 0.04957052 0.08169883 0.08724611 7.549536e-02 0.039440174
## LE2 0.01006307 0.12841183 0.09457992 2.747772e-02 0.002943328
## LE3 0.05278894 0.03158038 0.07019048 2.622030e-01 0.246744970
## LE4 0.30161978 0.03494590 0.08382893 4.458162e-15 0.009809012
## LE5 0.17506202 0.19692135 0.14271831 1.226647e-01 0.137689497
## LE6 0.09394966 0.09523241 0.13293934 2.158494e-01 0.155547222
- A heatmap of cell state abundances across the samples assigned to ecotypes. Rows correspond to the cell states forming ecotypes, while columns correspond to the samples assigned to ecotypes:
knitr::include_graphics("RecoveryOutput/bulk_lymphoma_data/Ecotypes/heatmap_assigned_samples_viridis.png")EcoTyper comes pre-loaded with the resources necessary for the reference-guided recovery of cell states and ecotypes previously defined in carcinoma and lymphoma, in user-provided scRNA-seq data.
In this tutorial, we illustrate how EcoTyper can be used to recover the cell states and ecotypes, that we defined across carcinomas and in diffuse large B cell lymphoma (DLBCL), in a downsampled version of a scRNA-seq dataset from colorectal cancer specimens, and a downsampled version of a scRNA-seq dataset from lymphoma specimens, respectively. Plese note that the recovery procedure described in this tutorial can also be applied on user-defined cell states and ecotypes, derived as described in Tutorials 4-6.
In this section we illustrate how carcinoma cell states can be recovered
in a scRNA-seq dataset from colorectal cancer specimens. The expression
data used in this tutorial can be found in
example_data/scRNA_CRC_data.txt, and its corresponding sample
annotation in example_data/scRNA_CRC_annotation.txt.
The script used to perform recovery in scRNA-seq data is
EcoTyper_recovery_scRNA.R:
Rscript EcoTyper_recovery_scRNA.R -h## usage: EcoTyper_recovery_scRNA.R [-d <character>] [-m <PATH>] [-a <PATH>]
## [-c <character>] [-z <bool>] [-s <integer>]
## [-t <integer>] [-o <PATH>] [-h]
##
## Arguments:
## -d <character>, --discovery <character>
## The name of the discovery dataset used to define cell
## states and ecotypes. Accepted values: 'Carcinoma' will
## recover the cell states and ecotypes defined across
## carcinomas, as described in the EcoTyper carcinoma
## paper, 'Lymphoma' will recover the cell states and
## ecotypes defined in diffuse large B cell lymphoma
## (DLBCL), as described in the EcoTyper lymphoma paper,
## '<MyDiscovery>' the value used in the field 'Discovery
## dataset name' of the config file used for running
## EcoTyper discovery ('EcoTyper_discovery.R') script.
## [default: 'Carcinoma']
## -m <PATH>, --matrix <PATH>
## Path to a tab-delimited file containing the input
## scRNA-seq expression matrix, with gene names on the
## first column and cell ids as column names [required].
## -a <PATH>, --annotation <PATH>
## Path to a tab-delimited annotation file containing the
## annotation of cells in the input matrix. This file
## should contain at least two columns, 'ID' with the
## same values as the columns of the expression matrix,
## and 'CellType' (case sensitive) which contains the
## cell type for each cell. These values are limited to
## the set of cell types analyzed in the discovery
## dataset. If the argument '-d' is set to 'Carcinoma',
## then the accepted values for column 'CellType' are:
## 'B.cells', 'CD4.T.cells', 'CD8.T.cells',
## 'Dendritic.cells', 'Endothelial.cells',
## 'Epithelial.cells', 'Fibroblasts', 'Mast.cells',
## 'Monocytes.and.Macrophages', 'NK.cells', 'PCs' and
## 'PMNs'. If the argument '-d' is set to 'Lymphoma',
## then the accepted values for column 'CellType' are:
## 'B.cells', 'Plasma.cells', 'T.cells.CD8',
## 'T.cells.CD4', 'T.cells.follicular.helper', 'Tregs',
## 'NK.cells', 'Monocytes.and.Macrophages',
## 'Dendritic.cells', 'Mast.cells', 'Neutrophils',
## 'Fibroblasts', 'Endothelial.cells'. All other values
## will be ignored for these two cases. Additionally,
## this file can contain any number of columns, that can
## be used for plotting color bars in the output heatmaps
## (see argument '-c'). [required]
## -c <character>, --columns <character>
## A comma-spearated list of column names from the
## annotation file to be plotted as color bar in the
## output heatmaps. [default: 'NULL']
## -z <bool>, --z-score <bool>
## A flag indicating whether the significance
## quantification procedure should be run. Note that this
## procedure might be slow, as the NMF model is applied
## 30 times on the same dataset. [default: 'FALSE']
## -s <integer>, --subsample <integer>
## An integer specifying the number of cells each cell
## type will be downsampled to. For values <50, no
## downsampling will be performed. [default: '-1' (no
## downsampling)]
## -t <integer>, --threads <integer>
## Number of threads. [default: '10']
## -o <PATH>, --output <PATH>
## Output directory path. [default: 'RecoveryOutput']
## -h, --help Print help message.
The script takes the following arguments:
-
-d/–discovery: The name of the discovery dataset used for defining cell states. By default, the only accepted values are Carcinoma and Lymphoma (case sensitive), which will recover the cell states that we already defined across carcinomas and in lymphoma, respectively. If the user defined cell states in their own data (Tutorial 4-6), the name of the discovery dataset is the value provided in the Discovery dataset name field of the configuration file used for running cell state discovery. For this tutorial, we set the name of the discovery dataset to Carcinoma.
-
-m/–matrix: Path to the input scRNA-seq matrix. The scRNA-seq expression matrix should be a tab-delimited file, with gene symbols on the first column and cells on the next columns. It should have cell identifiers (e.g. barcodes) as column names, and should be in TPM, CPM, FPKM or any other suitable count format. Gene symbols and cell identifiers should be unique. Moreover, we recommend that the column names do not contain special characters that are modified by the R function make.names, e.g. having digits at the beginning of the name or containing characters such as space, tab or -. The CRC cancer scRNA-seq data used in this tutorial looks as follows:
data = read.delim("example_data/scRNA_CRC_data.txt", nrow = 5)
head(data[,1:5])## Gene SMC01.T_AAAGATGCATGGATGG SMC01.T_AAAGTAGCAAGGACAC
## 1 A1BG 0 0
## 2 A1CF 0 0
## 3 A2M 0 0
## 4 A2ML1 0 0
## 5 A3GALT2 0 0
## SMC01.T_AAATGCCAGGATCGCA SMC01.T_AACTCTTCACAACGCC
## 1 0 0
## 2 0 0
## 3 0 0
## 4 0 0
## 5 0 0
- -a/–annotation: Path to a tab-delimited annotation file. This file should contain at least two columns: ID with the same values as the columns of the expression matrix, and CellType (case sensitive) which contains the cell type for each cell. The values in column CellType should indicate for each cell its cell type. These values are limited to the set of cell types analyzed in the discovery dataset. If the argument -d/–discovery is set to Carcinoma (as is the case for this tutorial), then the accepted values for column CellType are: ‘B.cells’, ‘CD4.T.cells’, ‘CD8.T.cells’, ‘Dendritic.cells’, ‘Endothelial.cells’, ‘Epithelial.cells’, ‘Fibroblasts’, ‘Mast.cells’, ‘Monocytes.and.Macrophages’, ‘NK.cells’, ‘PCs’ and ‘PMNs’. If the argument -d/–discovery is set to Lymphoma, then the accepted values for column CellType are: ‘B.cells’, ‘Plasma.cells’, ‘T.cells.CD8’, ‘T.cells.CD4’, ‘T.cells.follicular.helper’, ‘Tregs’, ‘NK.cells’, ‘Monocytes.and.Macrophages’, ‘Dendritic.cells’, ‘Mast.cells’, ‘Neutrophils’, ‘Fibroblasts’, ‘Endothelial.cells’. All other values will be ignored for these two cases. The annotation file can contain a column called Sample. If this column is present, the ecotype recovery will be performed, in addition to cell state recovery. Moreover, this file can contain any number of columns, that can be used for plotting color bars in the output heatmaps (see argument -c/–columns).
data = read.delim("example_data/scRNA_CRC_annotation.txt")
head(data)## Index Patient Class Sample Cell_type Cell_subtype
## 1 SMC01-T_AAAGATGCATGGATGG SMC01 Tumor SMC01-T Epithelial cells CMS2
## 2 SMC01-T_AAAGTAGCAAGGACAC SMC01 Tumor SMC01-T Epithelial cells CMS2
## 3 SMC01-T_AAATGCCAGGATCGCA SMC01 Tumor SMC01-T Epithelial cells CMS2
## 4 SMC01-T_AACTCTTCACAACGCC SMC01 Tumor SMC01-T Epithelial cells CMS2
## 5 SMC01-T_AACTTTCGTTCGGGCT SMC01 Tumor SMC01-T Epithelial cells CMS2
## 6 SMC01-T_AAGGTTCTCCAATGGT SMC01 Tumor SMC01-T Epithelial cells CMS2
## CellType ID Tissue
## 1 Epithelial.cells SMC01.T_AAAGATGCATGGATGG Tumor
## 2 Epithelial.cells SMC01.T_AAAGTAGCAAGGACAC Tumor
## 3 Epithelial.cells SMC01.T_AAATGCCAGGATCGCA Tumor
## 4 Epithelial.cells SMC01.T_AACTCTTCACAACGCC Tumor
## 5 Epithelial.cells SMC01.T_AACTTTCGTTCGGGCT Tumor
## 6 Epithelial.cells SMC01.T_AAGGTTCTCCAATGGT Tumor
-
-c/–columns: A comma-separated list of column names from the annotation file (see argument -a/–annotation) to be plotted as color bars in the output heatmaps. By default, the output heatmaps contain as color bar the cell state label each cell is assigned to. The column names indicated by this argument will be added to that color bar.
-
-z/–z-score: Flag indicating whether the significance quantification procedure should be run (default is FALSE). This procedure allows users to determine whether cell states are significantly recovered in a given dataset. Please note that this procedure can be very slow, as the NMF model is applied 30 times on the same dataset.
-
-s/–subsample: An integer specifying the number of cells each cell type will be downsampled to. For values <50, no downsampling will be performed. Default: -1 (no downsampling).
-
-t/–threads: Number of threads. Default: 10.
-
-o/–output: Output folder. The output folder will be created if it does not exist.
The command line for recovering the carcinoma cell states in the example scRNA-seq data is:
Rscript EcoTyper_recovery_scRNA.R -d Carcinoma -m example_data/scRNA_CRC_data.txt -a example_data/scRNA_CRC_annotation.txt -o RecoveryOutputThe outputs of this script include the following files, for each cell type provided:
- The assignment of single cells to states:
data = read.delim("RecoveryOutput/scRNA_CRC_data/Fibroblasts/state_assignment.txt")
head(data[,c("ID", "State")])## ID State
## 1 SMC01.T_TGCGCAGTCGGATGGA S01
## 2 SMC04.T_CACAAACTCTACTATC S01
## 3 SMC15.T_GCGCGATTCATAAAGG S01
## 4 SMC17.T_GTACGTAGTGACTACT S01
## 5 SMC20.T_CTAAGACCACTGTCGG S01
## 6 SMC20.T_GTTACAGTCGCGTTTC S01
- Two heatmaps: a heatmap representing the expression of cell state marker genes (see Tutorial 4 for more details) in the discovery dataset, and a heatmap with the expression of the same marker genes in the scRNA-seq dataset, smoothed to mitigate the impact of scRNA-seq dropout:
knitr::include_graphics("RecoveryOutput/scRNA_CRC_data/Fibroblasts/state_assignment_heatmap.png")- If the statistical significance quantification method is applied, the resulting z-scores for each cell state are output in the same directory:
#data = read.delim("RecoveryOutput/scRNA_CRC_data/Epithelial.cells/recovery_z_scores.txt")
#head(data[,c("State", "Z")])The output for ecotypes includes:
- The abundance (fraction) of each ecotype in each sample:
assign = read.delim("RecoveryOutput/scRNA_CRC_data/Ecotypes/ecotype_abundance.txt")
dim(assign)## [1] 10 33
head(assign[,1:5])## SMC01.N SMC01.T SMC02.N SMC02.T SMC03.N
## CE1 0.03013608 0.151493627 0.10825504 0.21601181 0.02930968
## CE2 0.00000000 0.009612861 0.00000000 0.02671883 0.00000000
## CE3 0.05199290 0.077968658 0.21188075 0.07300246 0.00000000
## CE4 0.01693878 0.043794663 0.03124202 0.00000000 0.00000000
## CE5 0.06285928 0.022531963 0.01115162 0.06406829 0.03197420
## CE6 0.16508230 0.059542004 0.24436913 0.01855854 0.43445664
- The assignment of samples to the carcinoma ecotype with the highest abundance. If the cell state fractions from the dominant ecotype are not significantly higher than the other cell state fractions in a given sample, the sample is considered unassigned and filtered out from this table. For more information about the sample filtering procedure please see the Ecotype discovery section of the EcoTyper paper methods:
discrete_assignments = read.delim("RecoveryOutput/scRNA_CRC_data/Ecotypes/ecotype_abundance.txt")
dim(discrete_assignments)## [1] 10 33
head(discrete_assignments[,1:5])## SMC01.N SMC01.T SMC02.N SMC02.T SMC03.N
## CE1 0.03013608 0.151493627 0.10825504 0.21601181 0.02930968
## CE2 0.00000000 0.009612861 0.00000000 0.02671883 0.00000000
## CE3 0.05199290 0.077968658 0.21188075 0.07300246 0.00000000
## CE4 0.01693878 0.043794663 0.03124202 0.00000000 0.00000000
## CE5 0.06285928 0.022531963 0.01115162 0.06406829 0.03197420
## CE6 0.16508230 0.059542004 0.24436913 0.01855854 0.43445664
- A heatmap of cell state abundances across the samples assigned to ecotypes. Rows correspond to the cell states forming ecotypes, while columns correspond to the samples assigned to ecotypes:
knitr::include_graphics("RecoveryOutput/scRNA_CRC_data/Ecotypes/heatmap_assigned_samples_viridis.png")In this section we illustrate how lymphoma cell states can be recovered
in the scRNA-seq dataset from lymphoma specimens. The expression data
used in this tutorial can be found in
example_data/scRNA_lymphoma_data.txt, and sample annotation in
example_data/scRNA_lymphoma_annotation.txt.
The script used to perform recovery in scRNA-seq data is called
EcoTyper_recovery_scRNA.R:
Rscript EcoTyper_recovery_scRNA.R -h## usage: EcoTyper_recovery_scRNA.R [-d <character>] [-m <PATH>] [-a <PATH>]
## [-c <character>] [-z <bool>] [-s <integer>]
## [-t <integer>] [-o <PATH>] [-h]
##
## Arguments:
## -d <character>, --discovery <character>
## The name of the discovery dataset used to define cell
## states and ecotypes. Accepted values: 'Carcinoma' will
## recover the cell states and ecotypes defined across
## carcinomas, as described in the EcoTyper carcinoma
## paper, 'Lymphoma' will recover the cell states and
## ecotypes defined in diffuse large B cell lymphoma
## (DLBCL), as described in the EcoTyper lymphoma paper,
## '<MyDiscovery>' the value used in the field 'Discovery
## dataset name' of the config file used for running
## EcoTyper discovery ('EcoTyper_discovery.R') script.
## [default: 'Carcinoma']
## -m <PATH>, --matrix <PATH>
## Path to a tab-delimited file containing the input
## scRNA-seq expression matrix, with gene names on the
## first column and cell ids as column names [required].
## -a <PATH>, --annotation <PATH>
## Path to a tab-delimited annotation file containing the
## annotation of cells in the input matrix. This file
## should contain at least two columns, 'ID' with the
## same values as the columns of the expression matrix,
## and 'CellType' (case sensitive) which contains the
## cell type for each cell. These values are limited to
## the set of cell types analyzed in the discovery
## dataset. If the argument '-d' is set to 'Carcinoma',
## then the accepted values for column 'CellType' are:
## 'B.cells', 'CD4.T.cells', 'CD8.T.cells',
## 'Dendritic.cells', 'Endothelial.cells',
## 'Epithelial.cells', 'Fibroblasts', 'Mast.cells',
## 'Monocytes.and.Macrophages', 'NK.cells', 'PCs' and
## 'PMNs'. If the argument '-d' is set to 'Lymphoma',
## then the accepted values for column 'CellType' are:
## 'B.cells', 'Plasma.cells', 'T.cells.CD8',
## 'T.cells.CD4', 'T.cells.follicular.helper', 'Tregs',
## 'NK.cells', 'Monocytes.and.Macrophages',
## 'Dendritic.cells', 'Mast.cells', 'Neutrophils',
## 'Fibroblasts', 'Endothelial.cells'. All other values
## will be ignored for these two cases. Additionally,
## this file can contain any number of columns, that can
## be used for plotting color bars in the output heatmaps
## (see argument '-c'). [required]
## -c <character>, --columns <character>
## A comma-spearated list of column names from the
## annotation file to be plotted as color bar in the
## output heatmaps. [default: 'NULL']
## -z <bool>, --z-score <bool>
## A flag indicating whether the significance
## quantification procedure should be run. Note that this
## procedure might be slow, as the NMF model is applied
## 30 times on the same dataset. [default: 'FALSE']
## -s <integer>, --subsample <integer>
## An integer specifying the number of cells each cell
## type will be downsampled to. For values <50, no
## downsampling will be performed. [default: '-1' (no
## downsampling)]
## -t <integer>, --threads <integer>
## Number of threads. [default: '10']
## -o <PATH>, --output <PATH>
## Output directory path. [default: 'RecoveryOutput']
## -h, --help Print help message.
The script takes the following arguments:
-
-d/–discovery: The name of the discovery dataset used for defining cell states. By default, the only accepted values are Carcinoma and Lymphoma (case sensitive), which will recover the cell states that we defined in carcinoma and lymphoma, respectively. If the user defined cell states in their own data (Tutorials 4-6), the name of the discovery dataset is the value provided in the ‘Discovery dataset name’ filed of the configuration file used for running EcoTyper discovery (‘EcoTyper_discovery_bulk.R’) script. In our tutorial, the name of the discovery dataset is Lymphoma.
-
-m/–matrix: Path to the input scRNA-seq matrix. The scRNA-seq expression matrix should be a tab-delimited file, with gene symbols on the first column and cells on the next columns. It should have cell identifiers (e.g. barcodes) as column names, and should be in TPM, CPM, FPKM or any other suitable count format. Gene symbols and cell identifiers should be unique. Moreover, we recommend that the column names do not contain special characters that are modified by the R function make.names, e.g. having digits at the beginning of the name or containing characters such as space, tab or -. The scRNA-seq data used in this tutorial looks as follows:
data = read.delim("example_data/scRNA_lymphoma_data.txt", nrow = 5)
head(data[,1:5])## Genes Cell_1 Cell_2 Cell_3 Cell_4
## 1 A1BG 0.0000 124.3626 0.0000 81.47967
## 2 A2M 0.0000 0.0000 0.0000 0.00000
## 3 A4GALT 0.0000 0.0000 0.0000 0.00000
## 4 AAAS 0.0000 0.0000 0.0000 0.00000
## 5 AACS 256.5418 0.0000 280.9778 0.00000
- -a/–annotation: Path to a tab-delimited annotation file. This file should contain at least two columns, ID with the same values as the columns of the expression matrix, and CellType which contains the cell type for each cell. The values in column CellType should be the same as the cell types analyzed in the discovery dataset. If the argument -d/–discovery is set to Lymphoma (as is the case for this tutorial), then the acceptable values for column CellType are: ‘B.cells’, ‘Dendritic.cells’, ‘Endothelial.cells’, ‘Fibroblasts’, ‘Mast.cells’, ‘Monocytes.and.Macrophages’, ‘Neutrophils’, ‘NK.cells’, ‘Plasma.cells’, ‘T.cells.CD4’, ‘T.cells.CD8’, ‘T.cells.follicular.helper’, and ‘Tregs’. All the other values will be ignored. The annotation file can contain a column called Sample. If this column is present, the ecotype recovery will be performed, in addition to cell state recovery. Moreover, this file can contain any number of columns, that can be used for plotting color bars in the output heatmaps (see argument -c/–columns).
data = read.delim("example_data/scRNA_lymphoma_annotation.txt")
head(data)## ID CellType Tissue
## 1 Cell_1 B.cells Tumor
## 2 Cell_2 B.cells Tumor
## 3 Cell_3 B.cells Tumor
## 4 Cell_4 B.cells Tumor
## 5 Cell_5 B.cells Tumor
## 6 Cell_6 B.cells Tumor
-
-c/–columns: A comma-separated list of column names from the annotation file (see argument -a/–annotation) to be plotted as color bars in the output heatmaps. By default, the output heatmaps contain as color bar the cell state label each cell is assigned to. The column names indicated by this argument will be added to that color bar.
-
-z/–z-score: Flag indicating whether the significance quantification procedure should be run (default is FALSE). This procedure allows users to determine whether cell states are significantly recovered in a given dataset. Please note that this procedure can be very slow, as the NMF model is applied 30 times on the same dataset.
-
-s/–subsample: An integer specifying the number of cells each cell type will be downsampled to. For values <50, no downsampling will be performed. Default: -1 (no downsampling).
-
-t/–threads: Number of threads. Default: 10.
-
-o/–output: Output folder. The output folder will be created if it does not exist.
The command line for recovering the lymphoma cell states in the example scRNA-seq data is:
Rscript EcoTyper_recovery_scRNA.R -d Lymphoma -m example_data/scRNA_lymphoma_data.txt -a example_data/scRNA_lymphoma_annotation.txt -o RecoveryOutput -c TissueThe outputs of this script include the following files, for each cell type provided:
- The assignment of single cells to states:
data = read.delim("RecoveryOutput/scRNA_lymphoma_data/B.cells/state_assignment.txt")
head(data[,c("ID", "State")])## ID State
## 1 Cell_2 S01
## 2 Cell_3 S01
## 3 Cell_4 S01
## 4 Cell_6 S01
## 5 Cell_8 S01
## 6 Cell_9 S01
- Two heatmaps: a heatmap representing the expression of cell state marker genes (see Tutorial 4 for more details) in the discovery dataset, and a heatmap with the expression of the same marker genes in the scRNA-seq dataset, smoothed to mitigate the impact of scRNA-seq dropout:
knitr::include_graphics("RecoveryOutput/scRNA_lymphoma_data/B.cells/state_assignment_heatmap.png")- If the statistical significance quantification method is applied, the resulting z-score for each cell state are output in the same directory:
#data = read.delim("RecoveryOutput/scRNA_lymphoma_data/B.cells/recovery_z_scores.txt")
#head(data[,c("State", "Z")])Since in this case the annotation file did not contain a column called Sample, ecotype recovery was not performed.
EcoTyper comes pre-loaded with the resources necessary for the reference-guided recovery of cell states and ecotypes previously defined in carcinoma and lymphoma, in user-provided expression data. The recovery procedure described in this tutorial can also be applied on user-defined cell states and ecotypes, derived as described in Tutorials 4-6.
Here we illustrate how one can perform cell state and ecotype recovery in Visium Spatial Gene Expression arrays from 10x Genomics. For this tutorial we recover cell states and ecotypes defined across carcinomas in whole transcriptome spatial transcriptomics data from breast cancer.
In order for EcoTyper to perform cell states and ecotypes recovery in Visium data, the following resources need to be available:
-
the filtered feature-barcode matrices
barcodes.tsv.gz,features.tsv.gzandmatrix.mtx.gz, in the format provided by 10x Genomics, and thetissue_positions_list.csvfile produced by the run summary images pipeline, containing the spatial position of barcodes. -
if the major cell populations expected in the system to be analyzed are recapitulated by the cell populations analyzed in the EcoTyper carcinoma paper (B cells, CD4 T cells, CD8 T cells, dendritic cells, endothelial cells, epithelial cells, fibroblasts, mast cells, monocytes/macrophages, NK cells, plasma cells, neutrophils), or the EcoTyper lymphoma paper (B cells, CD4 T cells, CD8 T cells, follicular helper T cells, Tregs, dendritic cells, endothelial cells, fibroblasts, mast cells, monocytes/macrophages, NK cells, plasma cells, neutrophils), then the user needs:
-
Docker
-
Docker containers for CIBERSORTx Fractions and CIBERSORTx HiRes modules, both of which can be obtained from the CIBERSORTx website. Please follow the instructions from the website to install them.
-
A token required for running the docker containers, which can also be obtained from the CIBERSORTx website.
-
-
if the major cell populations expected in the system to be analyzed are not recapitulated by the cell populations analyzed in the EcoTyper carcinoma paper (B cells, CD4 T cells, CD8 T cells, dendritic cells, endothelial cells, epithelial cells, fibroblasts, mast cells, monocytes/macrophages, NK cells, plasma cells, neutrophils), or the EcoTyper lymphoma paper (B cells, CD4 T cells, CD8 T cells, follicular helper T cells, Tregs, dendritic cells, endothelial cells, fibroblasts, mast cells, monocytes/macrophages, NK cells, plasma cells, neutrophils), then the user needs to provide their own cell type proportion estimations for these populations (see more details below).
The script that does cell type and ecotype discovery is:
Rscript EcoTyper_recovery_visium.R -h## usage: EcoTyper_recovery_visium.R [-c <PATH>] [-h]
##
## Arguments:
## -c <PATH>, --config <PATH>
## Path to the config files [required].
## -h, --help Print help message.
This script takes as input file a configuration file in
YAML format. The configuration file for this
tutorial is available in config_recovery_visium.yml:
default :
Input :
Discovery dataset name : "Carcinoma"
Recovery dataset name : "VisiumBreast"
Input Visium directory : "example_data/VisiumBreast"
#Path to a file containing the precomputed cell fractions for the visium array
Recovery cell type fractions : "NULL"
Malignant cell of origin : "Epithelial.cells"
CIBERSORTx username : "<Please use your username from the CIBERSORTx website>"
CIBERSORTx token : "<Please obtain a token from the CIBERSORTx website>"
Output :
Output folder : "VisiumOutput"
Pipeline settings :
Number of threads : 10The configuration file has three sections, Input, Pipeline settings, and Output. We next will describe the expected content in each of these sections, and instruct the user how to set the appropriate settings in their applications.
The Input section contains settings regarding the input data.
Discovery dataset name : "Carcinoma"Discovery dataset name should contain the name of the discovery dataset used for defining cell states. By default, the only accepted values are Carcinoma and Lymphoma (case sensitive), which will recover the cell states that we defined across carcinomas and in lymphoma, respectively. If the user defined cell states in their own data (Tutorials 4-6), the name of the discovery dataset is the value provided in the Discovery dataset name field of the configuration file used for running discovery. For this tutorial, we set the name of the discovery dataset to Carcinoma.
Recovery dataset name : "VisiumBreast"Recovery dataset name is the identifier used by EcoTyper to internally save and retrieve the information about the cell states/ecotypes abundances. Any value that contains alphanumeric characters and ’_’ is accepted for this field.
Input Visium directory : "example_data/VisiumBreast"There are 4 input files needed for recovery on the visium data:
list.files("example_data/VisiumBreast")## [1] "barcodes.tsv.gz" "features.tsv.gz"
## [3] "matrix.mtx.gz" "tissue_positions_list.csv"
The filtered feature-barcode matrices barcodes.tsv.gz,
features.tsv.gz and matrix.mtx.gz, in the format provided by 10x
Genomics,
and the tissue_positions_list.csv file produced by the run summary
images
pipeline,
containing the spatial position of barcodes.
Recovery cell type fractions : "NULL"Recovery cell type fractions should contain the path to a file containing the cell type fraction estimations for each spot on the visium array. This field is ignored when the discovery dataset is Carcinoma or Lymphoma or when the discovery has been performed as described in Tutorial 4, using Carcinoma_Fractions or Lymphoma_Fractions. It is only used when users provided their own cell type fractions for deriving cell states and ecotypes in Tutorial 4. In this case, the user needs to provide a path to a tab-delimited file for this field. The file should contain in the first column the same sample names used as column names in the input expression matrix, and in the next columns, the cell type fractions for the same cell populations used for discovering cell states and ecotypes. These fractions should sum up to 1 for each row. An example of such a file is provided in:
data = read.delim("example_data/visium_fractions_example.txt", nrow = 5)
dim(data)## [1] 5 13
data## Mixture Fibroblasts Endothelial.cells Epithelial.cells B.cells
## 1 AAACAAGTATCTCCCA.1 0.3747796 0.016948078 0.2164860 0.04116797
## 2 AAACACCAATAACTGC.1 0.1231510 0.028426736 0.6737582 0.02209104
## 3 AAACAGAGCGACTCCT.1 0.2383718 0.085296697 0.3124031 0.03159104
## 4 AAACAGGGTCTATATT.1 0.1178922 0.053757339 0.1128586 0.11847960
## 5 AAACAGTGTTCCTGGG.1 0.3699561 0.005238928 0.5008316 0.01311040
## CD4.T.cells CD8.T.cells Dendritic.cells Mast.cells Monocytes.and.Macrophages
## 1 0.10447645 0.033647446 0.016196773 0.021842932 0.06183330
## 2 0.04376232 0.025219723 0.006647209 0.008375436 0.03087553
## 3 0.04581258 0.028235504 0.025698640 0.020992271 0.04386487
## 4 0.11018235 0.154411312 0.004780762 0.013200087 0.09278191
## 5 0.02826441 0.007037966 0.005238555 0.006979820 0.02851720
## NK.cells PCs PMNs
## 1 0.030228865 0.06911276 0.013279833
## 2 0.006960189 0.02580987 0.004922716
## 3 0.020617429 0.13918007 0.007936014
## 4 0.000000000 0.21863726 0.003018626
## 5 0.007854087 0.02484208 0.002128834
Since in this tutorial we use the Carcinoma dataset as the discovery dataset, this field is not required. However, if it needs to be provided, it can be set as follows:
Recovery cell type fractions : "example_data/visium_fractions_example.txt"Malignant cell of origin : "Epithelial.cells"The cell of origin population for the cancer type being analyzed, amongst the cell types used for discovery. This field is used for plotting a gray background in the resulting output plot, with the intensity of gray depicting the abundance of the cell of origin population in each spot. It is not used when the discovery dataset is Carcinoma or Lymphoma or when the discovery has been performed as described in Tutorials 4-6, using Carcinoma_Fractions or Lymphoma_Fractions. In these cases, the malignant cells are automatically considered to be originating from Epithelial.cells or B.cells, respectively. Otherwise, this field needs to contain a column name in the file provided in Recovery cell type fractions field, corresponding to the appropriate cell type of origin.
CIBERSORTx username : "<Please use your username from the CIBERSORTx website>"
CIBERSORTx token : "<Please obtain a token from the CIBERSORTx website>"The fields CIBERSORTx username and CIBERSORTx token should contain the username on the CIBERSORTx website and the token necessary to run the CIBERSORTx source code. The token can be obtained from the CIBERSORTx website.
The Output section contains a single field, Output folder, which specifies the path where the final output will be saved. This folder will be created if it does not exist.
Output folder : "VisiumOutput"The last section, Pipeline settings, contains only one argument, the number of threads used for performing recovery:
Number of threads : 10After editing the configuration file (config_recovery_visium.yml), the
command line for recovering the cell states and ecotypes in Visium
Spatial Gene Expression data looks as illustrated below. Please note
that this script might take up to two hours to run on 10 threads. Also,
since CIBERSORTx is run on each spot, the memory requirements might
exceed the memory available on a typical laptop. We recommend that this
tutorial is run on a server with >32GB of RAM.
Rscript EcoTyper_recovery_visium.R -c config_recovery_visium.ymlEcoTyper generates for each cell type the following outputs:
- Cell state abundances:
data = read.delim("VisiumOutput/VisiumBreast/state_abundances.txt")
dim(data)## [1] 3813 59
head(data[,1:10])## ID X Y Sample Malignant B.cells_S01 B.cells_S02
## 1 AAACAAGTATCTCCCA.1 50 102 VisiumBreast 0.2164860 0.93234 0.0000000
## 2 AAACACCAATAACTGC.1 59 19 VisiumBreast 0.6737582 0.00000 0.5003005
## 3 AAACAGAGCGACTCCT.1 14 94 VisiumBreast 0.3124031 0.00000 0.0000000
## 4 AAACAGGGTCTATATT.1 47 13 VisiumBreast 0.1128586 1.00000 0.0000000
## 5 AAACAGTGTTCCTGGG.1 73 43 VisiumBreast 0.5008316 0.00000 0.2969141
## 6 AAACATTTCCCGGATT.1 61 97 VisiumBreast 0.7553180 0.00000 0.3589368
## B.cells_S03 B.cells_S04 CD4.T.cells_S01
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 1
## 5 0 0 0
## 6 0 0 0
- Plots illustrating the cell state abundance across state from each cell type. The intensity of charcoal represents the cell state abundance. The intensity of gray represents the fraction of the cancer cell of origin population:
knitr::include_graphics("VisiumOutput/VisiumBreast/Fibroblasts_spatial_heatmaps.png")- Ecotype abundances:
data = read.delim("VisiumOutput/VisiumBreast/ecotype_abundances.txt")
dim(data)## [1] 3813 13
head(data[,1:10])## ID X Y Sample Malignant E1
## VisiumBreast.1 AAACAAGTATCTCCCA.1 50 102 VisiumBreast 0.1142685 0.0000000
## VisiumBreast.2 AAACACCAATAACTGC.1 59 19 VisiumBreast 0.7334114 0.3751726
## VisiumBreast.3 AAACAGAGCGACTCCT.1 14 94 VisiumBreast 0.2441395 0.0000000
## VisiumBreast.4 AAACAGGGTCTATATT.1 47 13 VisiumBreast 0.0000000 0.4173973
## VisiumBreast.5 AAACAGTGTTCCTGGG.1 73 43 VisiumBreast 0.4992702 0.2870171
## VisiumBreast.6 AAACATTTCCCGGATT.1 61 97 VisiumBreast 0.8438427 0.1124896
## E2 E3 E4 E5
## VisiumBreast.1 0.0000000 0.0000000 0.5014347 0.3590839
## VisiumBreast.2 0.9696322 0.0000000 0.0000000 0.4942756
## VisiumBreast.3 0.0000000 0.0000000 0.0000000 0.0000000
## VisiumBreast.4 0.6267933 0.0000000 0.0000000 0.6267933
## VisiumBreast.5 0.1185850 0.0000000 0.5014347 0.0000000
## VisiumBreast.6 0.0000000 0.1890679 0.0000000 0.3792658
- Plots illustrating the ecotype abundances. The intensity of charcoal represents the cell state abundance. The intensity of gray represents the fraction of the cancer cell of origin population:
knitr::include_graphics("VisiumOutput/VisiumBreast/Ecotype_spatial_heatmaps.png")In this tutorial we illustrate how one can perform de novo
identification of cell states and ecotypes, starting from a bulk-tissue
expression matrix. For illustration purposes, we use as discovery
dataset a downsampled version of the TCGA samples from lung
adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), available
in example_data/bulk_lung_data.txt, together with the sample
annotation file example_data/bulk_lung_annotation.txt.
EcoTyper derives cell states and ecotypes in a sequence of steps:
-
Cell type fraction estimation: EcoTyper relies on cell abundance estimations of the major cell lineages expected to be present in the tissue analyzed, for each sample in the discovery dataset.
One way of estimating cell type abundances in bulk tissue specimens is by using CIBERSORTx Fractions module. CIBERSORTx Fractions leverages sets of barcode genes, termed signature matrix, to estimate cell fractions. Complete tutorials about how signature matrices can be derived are available on the CIBERSORTx website. In the EcoTyper carcinoma and lymphoma papers, we serially apply two signature matrices, to get a comprehensive representation of cell types typically found in these malignancies. We make these strategies automatically available in EcoTyper. If, however, the tissue/system being analyzed is expected to have different cell populations, then the user needs to estimate the appropriate fractions themselves (see details below). -
Cell type expression purification: To impute cell type-specific gene expression profiles from bulk tissue transcriptomes, EcoTyper employs CIBERSORTx HiRes module. CIBERSORTx HiRes takes as input the bulk expression matrix of the discovery dataset and the fractions of the cell populations obtained at step 1. It produces cell-type specific expression profiles, at single-sample resolution, for each cell population.
-
Cell state discovery: EcoTyper leverages nonnegative matrix factorization (NMF) to identify transcriptionally-defined cell states from expression profiles purified by CIBERSORTx HiRes (step 2). Given c cell types, let
be a
cell type-specific expression matrix for cell type
consisting of
rows (the number of genes) and
columns (the number of samples). The primary objective of NMF is to factorize
into two non-negative matrices: a
matrix,
, and a
matrix,
, where
is a user-specified rank (i.e., number of clusters). The basis matrix,
, encodes a representative expression level for each gene in each cell state. The mixture coefficients matrix
, scaled to sum to 1 across cell states, encodes the representation (relative abundance) of each cell state in each sample.
EcoTyper applies NMF on the top 1000 genes with highest relative dispersion across samples. If less than 1000 genes are available, all genes are selected. If less than 50 genes are imputed for a given cell type, that cell type is not used for cell state identification. Prior to NMF, each gene is scaled to mean 0 and unit variance. To satisfy the non-negativity requirement of NMF, cell type-specific expression matrices are individually processed using posneg transformation. This function converts an input expression matrixinto two matrices, one containing only positive values and the other containing only negative values with the sign inverted. These two matrices are subsequently concatenated to produce
.
For each cell type, EcoTyper applies NMF across a range of ranks (number of cell states), by default 2-20 states. For each rank, the NMF algorithm is applied multiple times (we recommend at least 50) with different starting seeds, for robustness. -
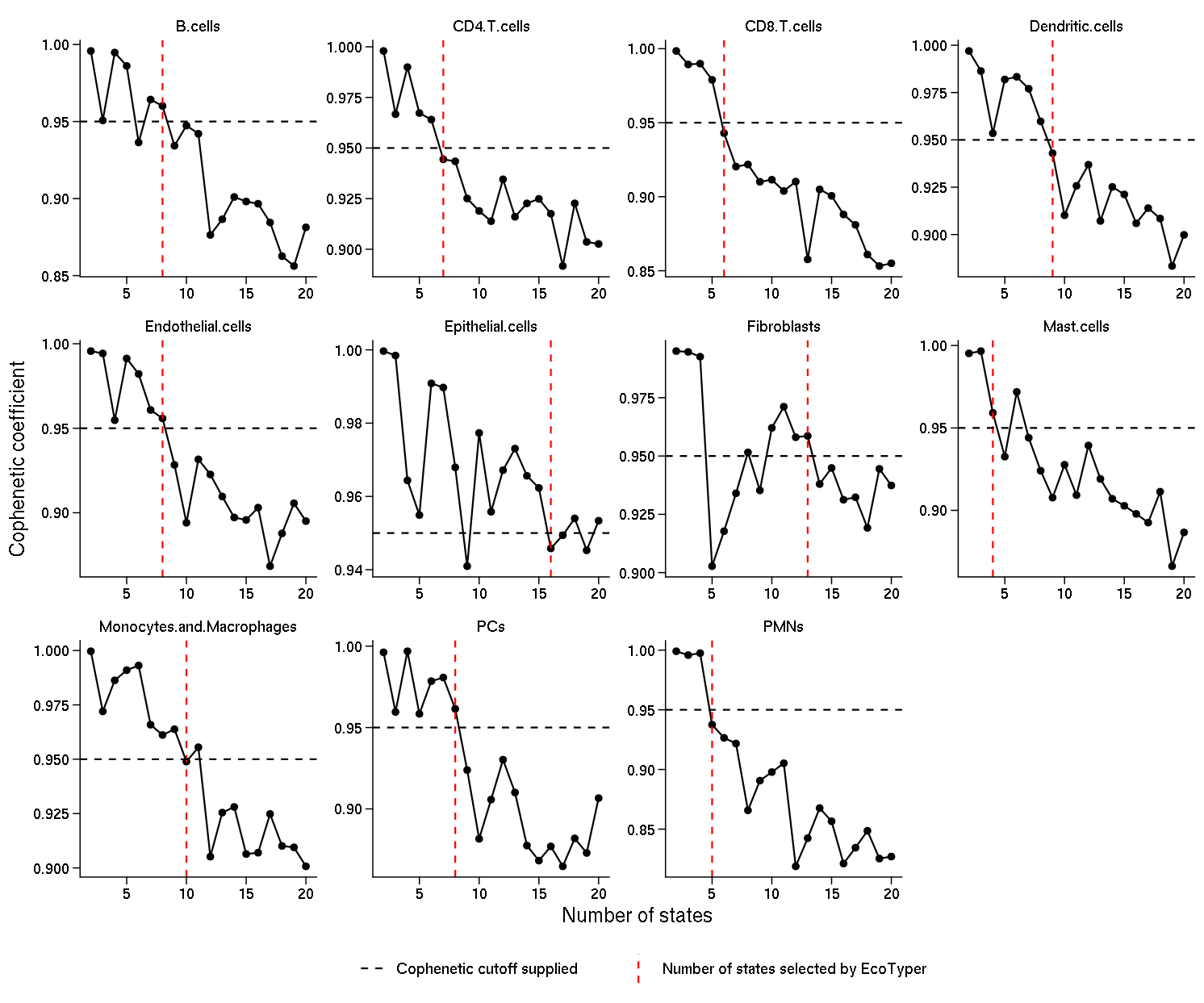
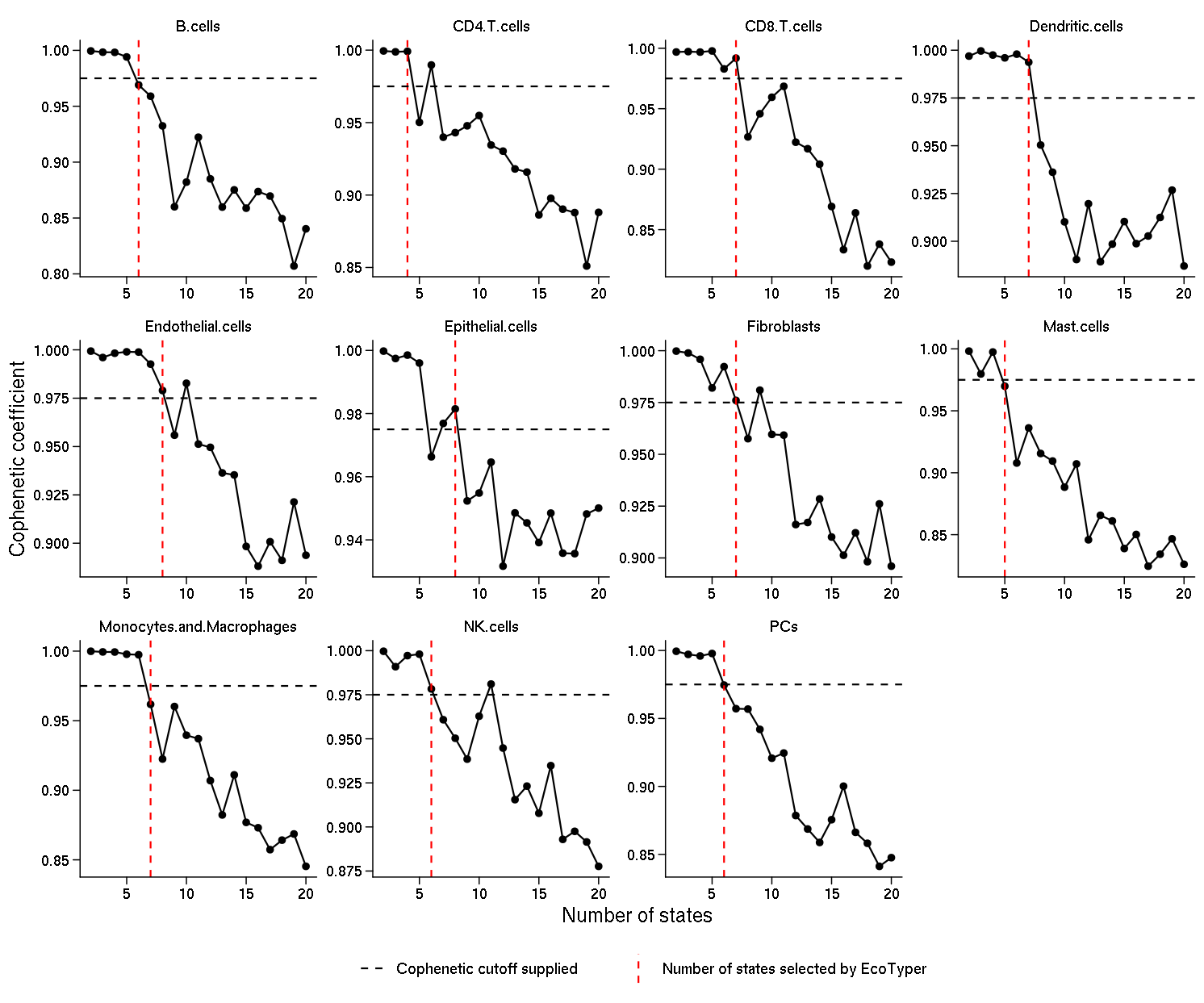
Choosing the number of cell states: Cluster (state) number selection is an important consideration in NMF applications. We found that previous approaches that rely on minimizing error measures (e.g., RMSE, KL divergence) or optimizing information-theoretic metrics either failed to converge or were dependent on the number of genes imputed. In contrast, the cophenetic coefficient quantifies the classification stability for a given rank (i.e., the number of clusters) and ranges from 0 to 1, with 1 being maximally stable. While the rank at which the cophenetic coefficient starts decreasing is typically selected, this approach is challenging to apply in situations where the cophenetic coefficient exhibits a multi-modal shape across ranks, as we found for some cell types. Therefore, we developed a heuristic approach more suitable for such settings. In each case, the rank was automatically chosen based on the cophenetic coefficient evaluated in the range 2–20 clusters (by default). Specifically, we determined the first occurrence in the interval 2–20 for which the cophenetic coefficient dropped below 0.95 (by default), having been above this level for at least two consecutive ranks. We then selected the rank immediately adjacent to this crossing point which was closest to 0.95 (by default).
-
Extracting cell state information: The NMF output resulting from step 4 is parsed and cell state information is extracted for the downstream analyses.
-
Cell state QC filter: Although posneg transformation is required to satisfy the non-negativity constraint of NMF following standardization, it can lead to the identification of spurious cell states driven by features with more negative values than positive ones. To combat this, we devised an adaptive false positive index (AFI), a novel index defined as the ratio between the sum of weights from the W matrix corresponding to the negative and positive features. EcoTyper automatically filters the states with
.
-
Advanced cell state QC filter: When the discovery dataset is comprised of multiple tumor types, we recommend using this advanced filter. This filter identifies poor-quality cell states using a dropout score, which flags states whose marker genes exhibit anomalously low variance and high expression across the discovery cohort, generally an artefact of CIBEROSRTx HiRes. To calculate the dropout score for each marker gene (i.e., genes with maximal log2 fold change in each state relative to other states within a given cell type), EcoTyper determines the maximum fraction of samples for which the gene has the same value. It also calculates the average log2 expression of the gene across samples. It averages each quantity, scaled to unit variance across states, within each state, converts them to z-scores, and removes states with a mean Z >1.96 (P < 0.05).
-
Ecotype (cellular community) discovery: Ecotypes or cellular communities are derived by identifying patterns of co-occurrence of cell states across samples. First, EcoTyper leverages the Jaccard index to quantify the degree of overlap between each pair of cell states across samples in the discovery cohort. Toward this end, it discretizes each cell state
into a binary vector
of length
, where
denotes the number of samples in the discovery cohort. Collectively, these vectors comprise binary matrix
, with same number of rows as cell states across cell types and
columns (samples). Given sample
, if state
is the most abundant state among all states in cell type
, EcoTyper sets
to 1; otherwise
. It then computes all pairwise Jaccard indices on the rows (states) in matrix
, yielding matrix
. Using the hypergeometric test, it evaluates the null hypothesis that any given pair of cell states
and
have no overlap. In cases where the hypergeometric p-value is >0.01, the Jaccard index for
is set to 0 (i.e., no overlap). To identify communities while accommodating outliers, the updated Jaccard matrix
is hierarchically clustered using average linkage with Euclidean distance (hclust in the R stats package). The optimal number of clusters is then determined via silhouette width maximization. Clusters with less than 3 cell states are eliminated from further analysis.
In order for EcoTyper to perform cell states and ecotypes discovery, the following resources need to be available:
-
docker containers for CIBERSORTx Fractions and CIBERSORTx HiRes modules, both of which can be obtained from the CIBERSORTx website. Please follow the instructions from the website to install them.
-
a token required for running the docker containers, which can also be obtained from the CIBERSORTx website.
-
a user-provided bulk tissue expression matrix (RNA-seq or microarray), on which the discovery will be performed (a discovery cohort). For this tutorial, we will use the example data in
example_data/bulk_lung_data.txt. -
if the major cell populations expected in the system to be analyzed are not recapitulated by the cell populations analyzed in the EcoTyper carcinoma paper (B cells, CD4 T cells, CD8 T cells, dendritic cells, endothelial cells, epithelial cells, fibroblasts, mast cells, monocytes/macrophages, NK cells, plasma cells, neutrophils), or the EcoTyper lymphoma paper (B cells, CD4 T cells, CD8 T cells, follicular helper T cells, Tregs, dendritic cells, endothelial cells, fibroblasts, mast cells, monocytes/macrophages, NK cells, plasma cells, neutrophils), then the user needs to provide their own cell type proportion estimations for these populations (see more details below).
-
optionally, a sample annotation file, such as the one provided in
example_data/bulk_lung_annotation.txt, can be supplied to EcoTyper. The information in this file can be used for heatmap plotting purposes, and also to instruct EcoTyper to find cell states/ecotypes common across different biological batches (e.g. tumor types), as detailed below.
The script that does cell type and ecotype discovery is:
Rscript EcoTyper_discovery_bulk.R -h## usage: EcoTyper_discovery_bulk.R [-c <PATH>] [-h]
##
## Arguments:
## -c <PATH>, --config <PATH>
## Path to the config files [required].
## -h, --help Print help message.
This script takes as input file a configuration file in
YAML format. The configuration file for this
tutorial is available in config_discovery_bulk.yml:
default :
Input :
Discovery dataset name : "MyDiscovery"
Expression matrix : "example_data/bulk_lung_data.txt"
#Possible values: "Carcinoma_Fractions", "Lymphoma_Fractions" or a path to a file containing the precomputed cell fractions
Cell type fractions : "Carcinoma_Fractions"
#Possible values: "RNA-seq", "Affymetrix", "Other"
Expression type : "RNA-seq"
#This field can also be set to "NULL"
Annotation file : "example_data/bulk_lung_annotation.txt"
#This field can also be set to "NULL"
Annotation file column to scale by : "Histology"
#This field can also be set to "NULL"
Annotation file column(s) to plot : ["Histology", "Tissue"]
CIBERSORTx username : "<Please use your username from the CIBERSORTx website>"
CIBERSORTx token : "<Please obtain a token from the CIBERSORTx website>"
Output :
Output folder : "DiscoveryOutput"
Pipeline settings :
#Pipeline steps:
# step 1 (cell type fraction estimation)
# step 2 (cell type expression purification)
# step 3 (cell state discovery)
# step 4 (choosing the number of cell states)
# step 5 (extracting cell state information)
# step 6 (cell state QC filter)
# step 7 (advanced cell state QC filter)
# step 8 (ecotype discovery)
Pipeline steps to skip : [7] # by default, step 7 is skipped
Number of threads : 10
Number of NMF restarts : 5
Maximum number of states per cell type : 20
Cophenetic coefficient cutoff : 0.95The configuration file has three sections, Input, Output and Pipeline settings. We next will describe the expected content in each of these three sections, and instruct the user how to set the appropriate settings in their applications.
The Input section contains settings regarding the input data.
Discovery dataset name is the identifier used by EcoTyper to
internally save and retrieve the information about the cell
states/ecotypes defined on this discovery dataset. It is also the name
to be provided to the -d/–discovery argument of scripts
EcoTyper_recovery_scRNA.R and EcoTyper_recovery_bulk.R, when
performing cell state/ecotypes recovery. Any value that contains
alphanumeric characters and ’_’ is accepted for this field.
Discovery dataset name : "MyDiscovery"Expression matrix : "example_data/bulk_lung_data.txt"Expression matrix field should contain the path to a tab-delimited file containing the expression data, with genes as rows and samples as columns. The expression matrix should be in the TPM or FPKM space for bulk RNA-seq and non-logarithmic (exponential) space for microarrays. It should have gene symbols on the first column and gene counts for each sample on the next columns. Column (sample) names should be unique. Also, we recommend that the column names do not contain special characters that are modified by the R function make.names, e.g. having digits at the beginning of the name or containing characters such as space, tab or -:
The expected format for the expression matrix is:
data = read.delim("example_data/bulk_lung_data.txt", nrow = 5)
dim(data)## [1] 5 251
head(data[,1:5])## Gene TCGA.37.A5EN.01A.21R.A26W.07 TCGA.37.4133.01A.01R.1100.07
## 1 A1BG 18.6400165 18.196602709
## 2 A1CF 0.0338368 0.002095014
## 3 A2M 54.1463351 35.714991125
## 4 A2ML1 4.9953315 2.383752067
## 5 A3GALT2 0.0438606 0.000000000
## TCGA.77.7465.01A.11R.2045.07 TCGA.34.5240.01A.01R.1443.07
## 1 24.83635354 23.579201761
## 2 0.02301987 0.004186634
## 3 80.63633736 86.804257397
## 4 4.08688641 3.015307103
## 5 0.00000000 0.000000000
Cell type fractions field instructs EcoTyper on how to compute cell type fractions on the discovery dataset:
#Possible values: "Carcinoma_Fractions", "Lymphoma_Fractions" or a path to a file containing the precomputed cell fractions
Cell type fractions : "Carcinoma_Fractions"If the major cell populations expected in the user-provided discovery dataset are recapitulated by the cell populations analyzed in the EcoTyper carcinoma paper (B cells, CD4 T cells, CD8 T cells, dendritic cells, endothelial cells, epithelial cells, fibroblasts, mast cells, monocytes/macrophages, NK cells, plasma cells and neutrophils), then this field can be set to Carcinoma_Fractions (case sensitive), and EcoTyper will automatically estimate fractions for these populations, in step 1 of the workflow. Similarly, if the cell populations analyzed in the EcoTyper lymphoma paper (B cells , CD4 T cells, CD8 T cells, follicular helper T cells, Tregs, dendritic cells, endothelial cells, fibroblasts, mast cells, monocytes/macrophages, NK cells, plasma cells and neutrophils) are appropriate, then the user can set this field to Lymphoma_Fractions and cell fractions will be automatically calculated.
In each of these cases the fractions are being estimated by serially
applying two signature matrices on the discovery dataset. The first
signature matrix, denoted TR4, is available in
utils/signature_matrices/TR4/TR4. TR4 was obtained from FACS-sorted
profiles of epithelial cells (EPCAM+), fibroblasts (CD10+), endothelial
cells (CD31+) and immune cells (CD45+), obtained from lung cancer
specimens (Newman et al., Nature Biotechnology
2019). The second
signature matrix, LM22, available in
utils/signature_matrices/LM22/LM22, was published with Newman et al.,
Nature Methods 2015, and is
able to deconvolve 22 immune subsets. In the EcoTyper carcinoma paper,
we first collapse the fractions for 22 subsets to obtain the
representation of the 9 major cell types (B cells, plasma cells, CD4 T
cells, CD8 T cells, NK cells, monocytes/macrophages, dendritic cells,
mast cells, and neutrophils). We then replace the TR4 immune cell
fractions with the fractions of the 9 cell lineages. This way we obtain
cell abundance estimations for 12 cell populations used in that paper.
An analogous process is used to obtain the lymphoma fractions.
If neither of these cases apply, the user needs to provide a path to a tab-delimited file containing the cell type proportion estimations for the expected populations. The file should contain in the first column the same sample names used for column names in the input expression matrix, and in the next columns, the cell type fractions for each cell population. These fractions should sum up to 1 for each row. An example of such a file is provided in:
data = read.delim("example_data/bulk_fractions_example.txt", nrow = 5)
dim(data)## [1] 5 13
data## Mixture Fibroblasts Endothelial.cells Epithelial.cells
## 1 TCGA.05.4249.01A.01R.1107.07 0.04016289 0.014982782 0.7054344
## 2 TCGA.05.4397.01A.01R.1206.07 0.02267369 0.009185669 0.6347924
## 3 TCGA.05.4398.01A.01R.1206.07 0.07151019 0.012282375 0.4041580
## 4 TCGA.05.4410.01A.21R.1858.07 0.03624798 0.009184835 0.6380211
## 5 TCGA.05.5425.01A.02R.1628.07 0.02596008 0.016036916 0.5820677
## B.cells CD4.T.cells CD8.T.cells Dendritic.cells Mast.cells
## 1 0.007229603 0.03216195 0.009618405 0.040271214 0.037739587
## 2 0.008658565 0.03586357 0.018784902 0.017186200 0.010598134
## 3 0.008732240 0.11555398 0.031922166 0.017226963 0.009799286
## 4 0.010793538 0.04992778 0.042050615 0.001396051 0.006484677
## 5 0.010044434 0.06866075 0.055325719 0.009598804 0.007838445
## Monocytes.and.Macrophages NK.cells PCs PMNs
## 1 0.08498321 0.003858362 0.02000494 0.003552648
## 2 0.20940083 0.012661304 0.01555575 0.004638961
## 3 0.25249613 0.013547285 0.05768268 0.005088728
## 4 0.11240034 0.008568742 0.08322750 0.001696815
## 5 0.17408990 0.012115993 0.03057943 0.007681873
This path can provided in the configuration file as follows:
#Possible values: "Carcinoma_Fractions", "Lymphoma_Fractions" or a path to a file containing the precomputed cell fractions
Cell type fractions : "example_data/bulk_fractions_example.txt" #Possible values: "RNA-seq", "Affymetrix", "Other"
Expression type : "RNA-seq"Expression type field specifies the platform used to generate the data provided in the expression matrix. The accepted values are RNA-seq for bulk RNA-seq data, Affymetrix for data profiled using Affymetrix microarray platforms, and Other for data from non-Affymetrix microarray platform. This argument is relevant only if the cell type fractions are being estimated automatically by EcoTyper (i.e. values Carcinoma_Fractions or Lymphoma_Fractions are being provided in the field Cell type fractions of the configuration file, as described above). Based on this field, EcoTyper determines the appropriate parameters for the CIBERSORTx fractions module, when estimating cell type fractions using the TR4 and LM22 signatures (see above). If RNA-seq is provided, CIBERSORTx with no batch correction is applied on TR4 and with B-mode batch correction on LM22. If Affymetrix is provided CIBERSORTx fractions with B-mode batch correction is applied on TR4 and with no batch correction on LM22. If Other is provided, CIBERSORTx fractions with B-mode batch correction is applied on both signatures
Annotation file : "example_data/bulk_lung_annotation.txt"A path to an annotation file can be provided in the field Annotation file. If provided, this file should contain a column called ID with the same names as the columns of the expression matrix, and any number of additional columns. The additional columns can be used for defining sample batches (see Section Annotation file column to scale by below) and for plotting color bars in the heatmaps output (see Section Annotation file column(s) to plot below). If not provided, this field needs to be set to “NULL”. For the current example, the annotation file has the following format:
annotation = read.delim("example_data/bulk_lung_annotation.txt", nrow = 5)
dim(annotation)## [1] 5 6
head(annotation)## ID Tissue Histology Type OS_Time
## 1 TCGA.37.A5EN.01A.21R.A26W.07 Tumor LUSC Primary Solid Tumor 660
## 2 TCGA.37.4133.01A.01R.1100.07 Tumor LUSC Primary Solid Tumor 238
## 3 TCGA.77.7465.01A.11R.2045.07 Tumor LUSC Primary Solid Tumor 990
## 4 TCGA.34.5240.01A.01R.1443.07 Tumor LUSC Primary Solid Tumor 1541
## 5 TCGA.05.4249.01A.01R.1107.07 Tumor LUAD Primary Solid Tumor 1523
## OS_Status
## 1 0
## 2 0
## 3 0
## 4 0
## 5 0
Annotation file column to scale by : "Histology"In order to discover pan-carcinoma cell states and ecotypes in the EcoType carcinoma paper, we standardize genes to mean 0 and unit 1 within each tumor type (histology). Field Annotation file column to scale by allows users to specify a column name in the annotation file, by which the samples will be grouped when performing standardization. The example discovery dataset used in this tutorial has samples from lung adenocarcinoma and lung squamous cell carcinoma. Therefore, for this tutorial we will use the Histology column to perform standardization.
However, this is an analytical choice, depending on the purpose of the analysis. If the users are interested in defining cell states and ecotypes regardless of tumor type-specificity, this argument can be set to “NULL”. In this case, the standardization will be applied across all samples in the discovery cohort. The same will happen if the annotation file is not provided.
Annotation file column(s) to plot : ["Histology", "Tissue"]Annotation file column(s) to plot field specifies which columns in the annotation file will be used as color bar in the output heatmaps, in addition to the cell state label or ecotype label column, plotted by default.
CIBERSORTx username : "<Please use your username from the CIBERSORTx website>"
CIBERSORTx token : "<Please obtain a token from the CIBERSORTx website>"The fields CIBERSORTx username and CIBERSORTx token should contain the username on the CIBERSORTx website and the token necessary to run the CIBERSORTx source code. The token can be obtained from the CIBERSORTx website.
The Output section contains a single field, Output folder, which specifies the path where the final output will be saved. This folder will be created if it does not exist.
Output folder : "DiscoveryOutput"The last section, Pipeline settings, contains settings controlling how EcoTyper is run.
#Pipeline steps:
# step 1 (cell type fraction estimation)
# step 2 (cell type expression purification)
# step 3 (cell state discovery)
# step 4 (choosing the number of cell states)
# step 5 (extracting cell state information)
# step 6 (cell state QC filter)
# step 7 (advanced cell state QC filter)
# step 8 (ecotype discovery)
Pipeline steps to skip : [7] # by default, step 7 is skippedThe Pipeline steps to skip option allows user to skip some of the steps outlined in section Overview of the EcoTyper workflow for discovering cell states. Please note that this option is only intended for cases when the pipeline had already been run once, and small adjustments are made to the parameters. For example, if the Cophenetic coefficient cutoff used in step 3 needs adjusting, the user might want to skip steps 1-2 and re-run from step 3 onwards.
Number of threads : 10The number of threads EcoTyper will be run on.
Number of NMF restarts : 5The NMF approach used by EcoTyper (Brunet et al.), can give slightly different results, depending on the random initialization of the algorithm. To obtain a stable solution, NMF is generally run multiple times with different seeds, and the solution that best explains the discovery data is chosen. Additionally, the variation of NMF solutions across restarts with different seeds is quantified using Cophenetic coefficients and used in step 4 of EcoTyper for selecting the number of states. The parameter Number of NMF restarts specifies how many restarts with different seed should EcoTyper perform for each rank selection, in each cell type. Since this is a very time consuming process, in this example we only use 5. However, for publication-quality results, we recommend at least 50 restarts.
Maximum number of states per cell type : 20Maximum number of states per cell type specifies the upper end of the range for the number of states possible for each cell type. The lower end is 2.
Cophenetic coefficient cutoff : 0.95This field indicates the Cophenetic coefficient cutoff, in the range [0, 1], used for automatically determining the number of states in step 4. Lower values generally lead to more clusters being identified.
After editing the configuration file (config_discovery_bulk.yml), the
de novo discovery cell states and ecotypes can be run as is
illustrated below. Please note that this script might take up to two
hours to run on 10 threads. Also, although EcoTyper can be run on the
example data from this tutorial using a typical laptop (16GB memory), it
might not be the case for larger datasets. We recommend that cell type
and ecotype discovery is generally run on a server with >32GB of RAM.
Rscript EcoTyper_discovery_bulk.R -c config_discovery_bulk.ymlEcoTyper generates for each cell type the following outputs:
-
Plots displaying the Cophenetic coefficient calculated in step 4. The horizontal dotted line indicates the Cophenetic coefficient cutoff provided in the configuration file Cophenetic coefficient cutoff field. The vertical dotted red line indicates the number of states automatically selected based on the Cophenetic coefficient cutoff provided. We recommend that users inspect this file to make sure that the automatic selection provides sensible results. If the user wants to adjust the Cophenetic coefficient cutoff after inspecting this plot, they can rerun the discovery procedure skipping steps 1-3. Please note that:
- These plots indicate the number of states obtained before applying the filters for low-quality states in steps 6 and 7. Therefore, the final results will probably contain fewer states.
- The plots below might look slightly different when generated with R versions other than R/3.6.0. This is because some EcoTyper steps, including NMF algorithm initialization, depend on random number generation. Althoguh EcoTyper sets random seeds before each such step, different R version output different random numbers for the same seed. To mitigate this issue, we recommend at least 50 NMF restarts when running EcoTyper.
knitr::include_graphics("DiscoveryOutput/rank_plot.png")-
For each cell type, the following outputs, exemplified here for endothelial cells, are produced:
- Abundances of cell states remaining after the QC filters in steps 6 and 7 (if run), across samples in the discovery dataset:
data = read.delim("DiscoveryOutput/Endothelial.cells/state_abundances.txt")
dim(data)## [1] 4 250
head(data[,1:5])## TCGA.37.A5EN.01A.21R.A26W.07 TCGA.37.4133.01A.01R.1100.07
## S01 4.657038e-15 3.396931e-15
## S02 4.313475e-01 3.396931e-15
## S03 4.657038e-15 3.396931e-15
## S04 5.532795e-02 3.396931e-15
## TCGA.77.7465.01A.11R.2045.07 TCGA.34.5240.01A.01R.1443.07
## S01 4.011227e-15 3.955821e-15
## S02 2.750005e-01 8.143772e-02
## S03 4.011227e-15 3.955821e-15
## S04 4.011227e-15 1.748715e-03
## TCGA.05.4249.01A.01R.1107.07
## S01 4.256051e-15
## S02 4.256051e-15
## S03 1.137231e-01
## S04 8.575277e-01
- Assignment of samples in the discovery dataset to the cell state with the highest abundance. Only samples assigned to the cell states remaining after the QC filters in steps 6 and 7 (if run) are included. The remaining ones are considered unassigned and removed from this table:
data = read.delim("DiscoveryOutput/Endothelial.cells/state_assignment.txt")
dim(data)## [1] 131 3
head(data)## ID State InitialState
## 31 TCGA.55.6983.11A.01R.1949.07 S01 IS02
## 32 TCGA.44.6776.11A.01R.1858.07 S01 IS02
## 33 TCGA.77.7335.11A.01R.2045.07 S01 IS02
## 34 TCGA.38.A44F.01A.11R.A24H.07 S01 IS02
## 35 TCGA.77.7138.11A.01R.2045.07 S01 IS02
## 36 TCGA.44.6778.11A.01R.1858.07 S01 IS02
- A heatmap illustrating the expression of genes used for cell state discovery, that have the highest fold-change in one of the cell states remaining after the QC filters in steps 6 and 7 (if run). In the current example, the heatmap includes in the top color bar two rows corresponding to Tissue and Histology, that have been provided in configuration file field Annotation file column(s) to plot, in addition to cell state labels always plotted:
knitr::include_graphics("DiscoveryOutput/Endothelial.cells/state_assignment_heatmap.png")The ecotype output files include:
- The cell state composition of each ecotype (the set of cell states making up each ecotype):
ecotypes = read.delim("DiscoveryOutput/Ecotypes/ecotypes.txt")
head(ecotypes[,c("CellType", "State", "Ecotype")])## CellType State Ecotype
## 1 B.cells S01 E1
## 2 Endothelial.cells S02 E1
## 3 Epithelial.cells S01 E1
## 4 Fibroblasts S07 E1
## 5 B.cells S03 E2
## 6 CD4.T.cells S02 E2
- The number of initial clusters obtained by clustering the Jaccard index matrix, selected using the average silhouette:
knitr::include_graphics("DiscoveryOutput/Ecotypes/nclusters_jaccard.png")- A heatmap of the Jaccard index matrix, after filtering ecotypes with less than 3 cell states:
knitr::include_graphics("DiscoveryOutput/Ecotypes/jaccard_matrix.png")- The abundance of each ecotype in each sample in the discovery dataset:
abundances = read.delim("DiscoveryOutput/Ecotypes/ecotype_abundance.txt")
dim(abundances)## [1] 7 250
head(abundances[,1:5])## TCGA.37.A5EN.01A.21R.A26W.07 TCGA.37.4133.01A.01R.1100.07
## E1 0.794531073 5.587512e-02
## E2 0.013996811 7.309504e-02
## E3 0.069214903 6.880715e-03
## E4 0.003306485 6.049462e-03
## E5 0.025837936 1.759492e-14
## E6 0.093112792 8.580997e-01
## TCGA.77.7465.01A.11R.2045.07 TCGA.34.5240.01A.01R.1443.07
## E1 4.508978e-01 9.485239e-01
## E2 3.120952e-01 2.957054e-03
## E3 7.323729e-15 1.835542e-03
## E4 8.909708e-15 1.377273e-02
## E5 1.645593e-01 7.296796e-15
## E6 7.244779e-02 3.291079e-02
## TCGA.05.4249.01A.01R.1107.07
## E1 4.493338e-09
## E2 1.180982e-01
## E3 5.358678e-01
## E4 1.804165e-01
## E5 8.538532e-02
## E6 1.303634e-15
- The assignment of samples in the discovery dataset to ecotypes. The samples not assigned to any ecotype are filtered out from this file:
assignments = read.delim("DiscoveryOutput/Ecotypes/ecotype_assignment.txt")
dim(assignments)## [1] 190 11
head(assignments[,1:5])## ID MaxEcotype
## TCGA.22.4613.01A.01R.1443.07 TCGA.22.4613.01A.01R.1443.07 E1
## TCGA.22.5471.01A.01R.1635.07 TCGA.22.5471.01A.01R.1635.07 E1
## TCGA.22.5473.01A.01R.1635.07 TCGA.22.5473.01A.01R.1635.07 E1
## TCGA.34.5240.01A.01R.1443.07 TCGA.34.5240.01A.01R.1443.07 E1
## TCGA.34.8456.01A.21R.2326.07 TCGA.34.8456.01A.21R.2326.07 E1
## TCGA.37.A5EM.01A.21R.A27Q.07 TCGA.37.A5EM.01A.21R.A27Q.07 E1
## AssignmentP AssignmentQ AssignedToEcotypeStates
## TCGA.22.4613.01A.01R.1443.07 0.006237509 0.02267708 TRUE
## TCGA.22.5471.01A.01R.1635.07 0.011589892 0.03408792 TRUE
## TCGA.22.5473.01A.01R.1635.07 0.173391227 0.21892832 TRUE
## TCGA.34.5240.01A.01R.1443.07 0.073719200 0.12710207 TRUE
## TCGA.34.8456.01A.21R.2326.07 0.002977868 0.01404655 TRUE
## TCGA.37.A5EM.01A.21R.A27Q.07 0.027084554 0.06573921 TRUE
- A heatmap of cell state fractions across the samples assigned to ecotypes:
knitr::include_graphics("DiscoveryOutput/Ecotypes/heatmap_assigned_samples_viridis.png")In this tutorial we illustrate how one can perform de novo
identification of cell states and ecotypes, starting from a
scRNA-seq expression matrix. For illustration purposes, we use as
discovery dataset a downsampled version of the scRNA-seq from colorectal
cancer, available in example_data/scRNA_CRC_data.txt, together with
the sample annotation file example_data/scRNA_CRC_annotation.txt.
EcoTyper derives cell states and ecotypes from scRNA-seq data in a sequence of steps:
-
Extract cell type specific genes: The removal of genes that are not specifically expressed in a given cell type, is an important consideration for reducing the likelihood of identifying spurious cell states. Ecotyper applies by default a filter for non-cell type specific genes, before performing cell state discovery in scRNA-seq data. Specifically, it performs a differential expression between cells from a given cell type and all other cell types combined. For computational efficency and balanced representation of cell types, only up to 500 randomly selected cells per cell type are used for this step. Genes with a Q-value > 0.05 (two-sided Wilcox test, with Benjamini-Hochberg correction for multiple hypothesis correction) are filtered out from each cell type. Of note, this filter is not necessary when discovering cell states in cell type specific profiles purified using CIBERSORTx high resolution (Tutorial 4). CIBERSORTx incorporates its own filter for genes without evidence of expression in a given cell type.
-
Cell state discovery on correlation matrices: EcoTyper leverages nonnegative matrix factorization (NMF) to identify transcriptionally-defined cell states from single cell expression profiles. However, NMF applied directly on scRNA-seq expression matrices may perform sub-optimally, since scRNA-seq data is generally sparse. Therefore, EcoTyper first applies NMF on the correlation matrix between each pair of cells from a given cell type. For computational efficency, EcoTyper only uses up to 2,500 randomly selected cells for this step.
To satisfy the non-negativity requirement of NMF, correlation matrices are individually processed using posneg transformation. This function converts a correlation matrixinto two matrices, one containing only positive values and the other containing only negative values with the sign inverted. These two matrices are subsequently concatenated to produce
.
For each cell type, EcoTyper applies NMF across a range of ranks (number of cell states), by default 2-20 states. For each rank, the NMF algorithm is applied multiple times (we recommend at least 50) with different starting seeds, for robustness. -
Choosing the number of cell states: Cluster (state) number selection is an important consideration in NMF applications. We found that previous approaches that rely on minimizing error measures (e.g., RMSE, KL divergence) or optimizing information-theoretic metrics either failed to converge or were dependent on the number of genes imputed. In contrast, the cophenetic coefficient quantifies the classification stability for a given rank (i.e., the number of clusters) and ranges from 0 to 1, with 1 being maximally stable. While the rank at which the cophenetic coefficient starts decreasing is typically selected, this approach is challenging to apply in situations where the cophenetic coefficient exhibits a multi-modal shape across ranks, as we found for some cell types. Therefore, we developed a heuristic approach more suitable for such settings. In each case, the rank was automatically chosen based on the cophenetic coefficient evaluated in the range 2–20 clusters (by default). Specifically, we determined the first occurrence in the interval 2–20 for which the cophenetic coefficient dropped below 0.95 (by default), having been above this level for at least two consecutive ranks. We then selected the rank immediately adjacent to this crossing point which was closest to 0.95 (by default).
-
Extracting cell state information: The NMF output resulting from step 2 is parsed and cell state information is extracted for the downstream analyses.
-
Cell state re-discovery on expression matrices: Following the identification of cell states on correlation matrices, EcoTyper performs differential expression to identify genes most highly associated with each cell state. The resulting markers are ranked by the fold-change in each state, and the top 1000 genes with the highest rank across cell states are selected for a new round of NMF. If less than 1000 genes are available, all genes are selected. Prior to NMF, each gene is scaled to mean 0 and unit variance. To satisfy the non-negativity requirement of NMF, cell type-specific expression matrices are individually processed using posneg transformation. This function converts an input expression matrix
into two matrices, one containing only positive values and the other containing only negative values with the sign inverted. These two matrices are subsequently concatenated to produce
.
For each cell type, EcoTyper only applies NMF for the rank selected in step 3. As before, the NMF algorithm is applied multiple times (we recommend at least 50) with different starting seeds, for robustness. -
Extracting cell state information: The NMF output resulting from step 5 is parsed and cell state information is extracted for the downstream analyses.
-
Cell state QC filter: Although posneg transformation is required to satisfy the non-negativity constraint of NMF following standardization, it can lead to the identification of spurious cell states driven by features with more negative values than positive ones. To combat this, we devised an adaptive false positive index (AFI), a novel index defined as the ratio between the sum of weights from the W matrix corresponding to the negative and positive features. EcoTyper automatically filters the states with
.
-
Ecotype (cellular community) discovery: Ecotypes or cellular communities are derived by identifying patterns of co-occurrence of cell states across samples. First, EcoTyper leverages the Jaccard index to quantify the degree of overlap between each pair of cell states across samples in the discovery cohort. Toward this end, it discretizes each cell state
into a binary vector
of length
, where
= the number of samples in the discovery cohort. Collectively, these vectors comprise binary matrix
, with same number of rows as cell states across cell types and
columns (samples). Given sample
, if state
is the most abundant state among all states in cell type
, EcoTyper sets
to 1; otherwise
. It then computes all pairwise Jaccard indices on the rows (states) in matrix
, yielding matrix
. Using the hypergeometric test, it evaluates the null hypothesis that any given pair of cell states
and
have no overlap. In cases where the hypergeometric p-value is >0.01, the Jaccard index for
is set to 0 (i.e., no overlap). To identify communities while accommodating outliers, the updated Jaccard matrix
is hierarchically clustered using average linkage with Euclidean distance (hclust in the R stats package). The optimal number of clusters is then determined via silhouette width maximization. Clusters with less than 3 cell states are eliminated from further analysis.
In order for EcoTyper to perform cell states and ecotypes discovery, the following resources need to be available:
-
a user-provided scRNA-seq expression matrix, on which the discovery will be performed (a discovery cohort). For this tutorial, we will use the example data in
example_data/scRNA_CRC_data.txt. -
a sample annotation file, such as the one provided in
example_data/scRNA_CRC_annotation.txt, with at least three columns: ID, CellType and Sample.
The script that does cell type and ecotype discovery is:
Rscript EcoTyper_discovery_scRNA.R -h## usage: EcoTyper_discovery_scRNA.R [-c <PATH>] [-h]
##
## Arguments:
## -c <PATH>, --config <PATH>
## Path to the config files [required].
## -h, --help Print help message.
This script takes as input file a configuration file in
YAML format. The configuration file for this
tutorial is available in config_discovery_scRNA.yml:
default :
Input :
Discovery dataset name : "discovery_scRNA_CRC"
Expression matrix : "example_data/scRNA_CRC_data.txt"
Annotation file : "example_data/scRNA_CRC_annotation.txt"
Annotation file column to scale by : NULL
Annotation file column(s) to plot : []
Output :
Output folder : "DiscoveryOutput_scRNA"
Pipeline settings :
#Pipeline steps:
# step 1 (extract cell type specific genes)
# step 2 (cell state discovery on correrlation matrices)
# step 3 (choosing the number of cell states)
# step 4 (extracting cell state information)
# step 5 (cell state re-discovery in expression matrices)
# step 6 (extracting information for re-discovered cell states)
# step 7 (cell state QC filter)
# step 8 (ecotype discovery)
Pipeline steps to skip : []
Filter non cell type specific genes : True
Number of threads : 10
Number of NMF restarts : 5
Maximum number of states per cell type : 20
Cophenetic coefficient cutoff : 0.95
#The p-value cutoff used for filtering non-significant overlaps in the jaccard matrix used for discovering ecotypes in step 8. Default: 1 (no filtering).
Jaccard matrix p-value cutoff : 1The configuration file has three sections, Input, Output and Pipeline settings. We next will describe the expected content in each of these three sections, and instruct the user how to set the appropriate settings in their applications.
The Input section contains settings regarding the input data.
Discovery dataset name is the identifier used by EcoTyper to
internally save and retrieve the information about the cell
states/ecotypes defined on this discovery dataset. It is also the name
to be provided to the -d/–discovery argument of scripts
EcoTyper_recovery_scRNA.R and EcoTyper_recovery_bulk.R, when
performing cell state/ecotypes recovery. Any value that contains
alphanumeric characters and ’_’ is accepted for this field.
Discovery dataset name : "discovery_scRNA_CRC"Expression matrix : "example_data/scRNA_CRC_data.txt"Expression matrix field should contain the path to a tab-delimited file containing the expression data, with genes as rows and cells as columns. The expression matrix should be in the TPM, CPM or other suitable normalized space. The users should perform their own quality control of the expression matrix before applying EcoTyper (e.g. to filter low-quality cells, doublets, etc.). However we do not recommend to pre-filter the matrix for variable genes, as EcoTyper performs an internal selection for genes that show cell-type specificity. The matrix should have gene symbols on the first column and gene counts for each cell on the next columns. Column (cells) names should be unique. Also, we recommend that the column names do not contain special characters that are modified by the R function make.names, e.g. having digits at the beginning of the name or containing characters such as space, tab or -:
The expected format for the expression matrix is:
data = read.delim("example_data/scRNA_CRC_data.txt", nrow = 5)
dim(data)## [1] 5 13781
head(data[,1:5])## Gene SMC01.T_AAAGATGCATGGATGG SMC01.T_AAAGTAGCAAGGACAC
## 1 A1BG 0 0
## 2 A1CF 0 0
## 3 A2M 0 0
## 4 A2ML1 0 0
## 5 A3GALT2 0 0
## SMC01.T_AAATGCCAGGATCGCA SMC01.T_AACTCTTCACAACGCC
## 1 0 0
## 2 0 0
## 3 0 0
## 4 0 0
## 5 0 0
Annotation file : "example_data/scRNA_CRC_annotation.txt"A path to an annotation file should be provided in the field Annotation file. This file should contain a column called ID with the same names (e.g. cell barcodes) as the columns of the expression matrix, a column called CellType indicating cell type for each cell, and a column called Sample indicating the sample identifier for each cell. The latter is used for ecotype discovery. This file can contain any number of additional columns. The additional columns can be used for defining sample batches (see Section Annotation file column to scale by below) and for plotting color bars in the heatmaps output (see Section Annotation file column(s) to plot below). For the current example, the annotation file has the following format:
annotation = read.delim("example_data/scRNA_CRC_annotation.txt", nrow = 5)
dim(annotation)## [1] 5 9
head(annotation)## Index Patient Class Sample Cell_type Cell_subtype
## 1 SMC01-T_AAAGATGCATGGATGG SMC01 Tumor SMC01-T Epithelial cells CMS2
## 2 SMC01-T_AAAGTAGCAAGGACAC SMC01 Tumor SMC01-T Epithelial cells CMS2
## 3 SMC01-T_AAATGCCAGGATCGCA SMC01 Tumor SMC01-T Epithelial cells CMS2
## 4 SMC01-T_AACTCTTCACAACGCC SMC01 Tumor SMC01-T Epithelial cells CMS2
## 5 SMC01-T_AACTTTCGTTCGGGCT SMC01 Tumor SMC01-T Epithelial cells CMS2
## CellType ID Tissue
## 1 Epithelial.cells SMC01.T_AAAGATGCATGGATGG Tumor
## 2 Epithelial.cells SMC01.T_AAAGTAGCAAGGACAC Tumor
## 3 Epithelial.cells SMC01.T_AAATGCCAGGATCGCA Tumor
## 4 Epithelial.cells SMC01.T_AACTCTTCACAACGCC Tumor
## 5 Epithelial.cells SMC01.T_AACTTTCGTTCGGGCT Tumor
Annotation file column to scale by : NULLIn order to discover pan-carcinoma cell states and ecotypes in the EcoType carcinoma paper, we standardize genes to mean 0 and unit variance within each tumor type (histology). Field Annotation file column to scale by allows users to specify a column name in the annotation file, by which the cells will be grouped when performing standardization. However, this is an analytical choice, depending on the purpose of the analysis. If the users are interested in defining cell states and ecotypes regardless of tumor type-specificity, this argument can be set to NULL. In this case, the standardization will be applied across all samples in the discovery cohort. The same will happen if the annotation file is not provided.
In the current example, this field is not used and therefore set to NULL.
Annotation file column(s) to plot : ["Histology", "Tissue"]Annotation file column(s) to plot field specifies which columns in the annotation file will be used as color bar in the output heatmaps, in addition to the cell state label column, plotted by default.
The Output section contains a single field, Output folder, which specifies the path where the final output will be saved. This folder will be created if it does not exist.
Output folder : "DiscoveryOutput_scRNA"The last section, Pipeline settings, contains settings controlling how EcoTyper is run.
#Pipeline steps:
# step 1 (extract cell type specific genes)
# step 2 (cell state discovery on correlation matrices)
# step 3 (choosing the number of cell states)
# step 4 (extracting cell state information)
# step 5 (cell state re-discovery in expression matrices)
# step 6 (extracting information for re-discovered cell states)
# step 7 (cell state QC filter)
# step 8 (ecotype discovery)The Pipeline steps to skip option allows user to skip some of the steps outlined in section Overview of the EcoTyper workflow for discovering cell states. Please note that this option is only intended for cases when the pipeline had already been run once, and small adjustments are made to the parameters. For example, if the Cophenetic coefficient cutoff used in step 3 needs adjusting, the user might want to skip steps 1-2 and re-run from step 3 onwards.
Filter non cell type specific genes : TrueFlag indicated whether to apply the filter for cell type specific genes in step 1, outlined in section Overview of the EcoTyper workflow for discovering cell states. For best results, we do recommend applying this filter.
Number of threads : 10The number of threads EcoTyper will be run on.
Number of NMF restarts : 5The NMF approach used by EcoTyper (Brunet et al.), can give slightly different results, depending on the random initialization of the algorithm. To obtain a stable solution, NMF is generally run multiple times with different seeds, and the solution that best explains the discovery data is chosen. Additionally, the variation of NMF solutions across restarts with different seeds is quantified using Cophenetic coefficients and used in step 4 of EcoTyper for selecting the number of states. The parameter Number of NMF restarts specifies how many restarts with different seed should EcoTyper perform for each rank selection, in each cell type. Since this is a very time consuming process, in this example we only use 5. However, for publication-quality results, we recommend at least 50 restarts.
Maximum number of states per cell type : 20Maximum number of states per cell type specifies the upper end of the range for the number of states possible for each cell type. The lower end is 2.
Cophenetic coefficient cutoff : 0.975This field indicates the Cophenetic coefficient cutoff, in the range [0, 1], used for automatically determining the number of states in step 4. Lower values generally lead to more clusters being identified. In this particular example, we set it to 0.975.
Jaccard matrix p-value cutoff : 1Ecotype identification on step 8 is performed by clustering a jaccard matrix that quantifies the sample overlap between each pair of states. Prior to performing ecotype identification, the jaccard matrix values corresponding to pairs of states for which the sample overlap is not significant are set to 0, in order to mitigate the noise introduced by spurious overlaps. The value provided in this field specifies the p-value cutoff above which the overlaps are considered non-significant. When the number of samples in the scRNA-seq dataset is small, such as in the current example, we recommend this filter is disabled (p-value cutoff = 1), to avoid over-filtering the jaccard matrix. However, we encourage users to set this cutoff to lower values (e.g. 0.05), if the discovery scRNA-seq dataset contains a number of samples large enough to reliably evaluate the significance of overlaps.
After editing the configuration file (config_discovery_scRNA.yml), the
de novo discovery cell states and ecotypes can be run as is
illustrated below. Please note that this script might take 24-48 hours
to run on 10 threads. Also, EcoTyper cannot be run on the example data
from this tutorial using a typical laptop (16GB memory). We recommend
that it is run on a server with >50-100GB of RAM or a high performance
cluster.
Rscript EcoTyper_discovery_scRNA.R -c config_discovery_scRNA.ymlEcoTyper generates for each cell type the following outputs:
-
Plots displaying the Cophenetic coefficient calculated in step 4. The horizontal dotted line indicates the Cophenetic coefficient cutoff provided in the configuration file Cophenetic coefficient cutoff field. The vertical dotted red line indicates the number of states automatically selected based on the Cophenetic coefficient cutoff provided. We recommend that users inspect this file to make sure that the automatic selection provides sensible results. If the user wants to adjust the Cophenetic coefficient cutoff after inspecting this plot, they can rerun the discovery procedure skipping steps 1-3. Please note that:
- These plots indicate the number of states obtained before applying the filters for low-quality states in steps 6 and 7. Therefore, the final results will probably contain fewer states.
- The plots below might look slightly different when generated with R versions other than R/3.6.0. This is because some EcoTyper steps, including NMF algorithm initialization, depend on random number generation. Althoguh EcoTyper sets random seeds before each such step, different R version output different random numbers for the same seed. To mitigate this issue, we recommend at least 50 NMF restarts when running EcoTyper.
knitr::include_graphics("DiscoveryOutput_scRNA/rank_plot.png")-
For each cell type, the following outputs, exemplified here for fibroblasts, are produced:
- Abundances of cell states remaining after the QC filters in steps 6 and 7 (if run), across samples in the discovery dataset:
data = read.delim("DiscoveryOutput_scRNA/Fibroblasts/state_abundances.txt")
dim(data)## [1] 5 1500
head(data[,1:5])## SMC01.T_AAAGTAGAGTGGTAGC SMC01.T_ACACCCTGTTGGTAAA SMC01.T_ACATCAGTCGCCTGAG
## S01 2.074345e-14 4.262766e-02 1.352784e-14
## S02 2.074345e-14 3.332167e-01 3.957569e-01
## S03 2.781436e-02 1.204917e-02 1.706085e-01
## S04 2.074345e-14 9.441681e-13 1.352784e-14
## S05 2.074345e-14 6.121064e-01 7.391235e-02
## SMC01.T_ACTATCTAGCTAGTCT SMC01.T_ACTGATGAGCACCGCT
## S01 5.529717e-02 1.022447e-14
## S02 2.081811e-14 1.805516e-01
## S03 3.519208e-02 6.787254e-01
## S04 2.081811e-14 1.022447e-14
## S05 2.081811e-14 7.112757e-02
- Assignment of samples in the discovery dataset to the cell state with the highest abundance. Only samples assigned to the cell states remaining after the QC filters in steps 6 and 7 (if run) are included. The remaining ones are considered unassigned and removed from this table:
data = read.delim("DiscoveryOutput_scRNA/Fibroblasts/state_assignment.txt")
dim(data)## [1] 899 3
head(data)## ID State InitialState
## 723 SMC15.T_CATCGAAGTGACCAAG S01 IS05
## 724 SMC18.T_CTTGGCTCAGTGACAG S01 IS05
## 725 SMC24.T_TACTTACAGCGCCTTG S01 IS05
## 726 SMC01.N_CACCAGGCAATAAGCA S01 IS05
## 727 SMC02.N_AGAGCTTTCTAACCGA S01 IS05
## 728 SMC02.N_ATAACGCCAATACGCT S01 IS05
- A heatmap illustrating the expression of genes used for cell state discovery, that have the highest fold-change in one of the cell states remaining after the QC filters in steps 6 and 7 (if run). In the current example, the heatmap includes in the top color bar two rows corresponding to Tissue and Histology, that have been provided in configuration file field Annotation file column(s) to plot, in addition to cell state labels always plotted:
knitr::include_graphics("DiscoveryOutput_scRNA/Fibroblasts/state_assignment_heatmap.png")The ecotype output files include:
- The cell state composition of each ecotype (the set of cell states making up each ecotype):
ecotypes = read.delim("DiscoveryOutput_scRNA/Ecotypes/ecotypes.txt")
head(ecotypes[,c("CellType", "State", "Ecotype")])## CellType State Ecotype
## 1 B.cells S02 E1
## 2 CD4.T.cells S02 E1
## 3 CD8.T.cells S01 E1
## 4 Dendritic.cells S03 E1
## 5 Fibroblasts S05 E1
## 6 Monocytes.and.Macrophages S03 E1
- The number of initial clusters obtained by clustering the Jaccard index matrix, selected using the average silhouette:
knitr::include_graphics("DiscoveryOutput_scRNA/Ecotypes/nclusters_jaccard.png")- A heatmap of the Jaccard index matrix, after filtering ecotypes with less than 3 cell states:
knitr::include_graphics("DiscoveryOutput_scRNA/Ecotypes/jaccard_matrix.png")- The abundance of each ecotype in each sample in the discovery dataset:
abundances = read.delim("DiscoveryOutput_scRNA/Ecotypes/ecotype_abundance.txt")
dim(abundances)## [1] 9 33
head(abundances[,1:5])## SMC01.N SMC01.T SMC02.N SMC02.T SMC03.N
## E1 0.34064095 0.07302366 0.20329837 0.02049678 0.27718758
## E2 0.06078240 0.17093342 0.02937202 0.10322721 0.05241208
## E3 0.02315383 0.34562878 0.01355632 0.36202739 0.01278497
## E4 0.13787420 0.12543986 0.14604672 0.16681631 0.06725426
## E5 0.16081886 0.10434607 0.28980392 0.11903111 0.14459666
## E6 0.00000000 0.07347385 0.03524642 0.14282270 0.00000000
- The assignment of samples in the discovery dataset to ecotypes. The samples not assigned to any ecotype are filtered out from this file:
assignments = read.delim("DiscoveryOutput_scRNA/Ecotypes/ecotype_assignment.txt")
dim(assignments)## [1] 32 6
head(assignments[,1:5])## ID MaxEcotype AssignmentP AssignmentQ AssignedToEcotypeStates
## SMC01-N SMC01-N E1 1.938649e-04 0.0012795085 TRUE
## SMC05-N SMC05-N E1 5.000404e-03 0.0183348142 TRUE
## SMC05-T SMC05-T E1 7.568441e-02 0.1541417608 TRUE
## SMC07-N SMC07-N E1 2.928585e-03 0.0138061877 TRUE
## SMC08-N SMC08-N E1 9.015769e-05 0.0007438009 TRUE
## SMC19-T SMC19-T E1 5.936002e-03 0.0195888071 TRUE
- A heatmap of cell state fractions across the samples assigned to ecotypes:
knitr::include_graphics("DiscoveryOutput_scRNA/Ecotypes/heatmap_assigned_samples_viridis.png")In this tutorial we illustrate how one can perform de novo identification of cell states and ecotypes, starting from cell-type specific expression matrices, obtained either through FACS-sorting cell populations of interest and then peforming bulk tissue expression profiling of each cell population, or by performing in silico purification, using CIBERSORTx or any other tool. For illustration purposes, we use the cell type specific profiles inferred by CIBERSORTx in Tutorial 4, based on a downsampled version of the TCGA samples from lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC).
EcoTyper derives cell states and ecotypes in a sequence of steps:
-
Extract cell type specific genes: The removal of genes that are not specifically expressed in a given cell type, is an important consideration for reducing the likelihood of identifying spurious cell states. Ecotyper applies by default a filter for non-cell type specific genes, before performing cell state discovery in pre-sorted data. Specifically, it performs a differential expression between cells from a given cell type and all other cell types combined. For computational efficency and balanced representation of cell types, only up to 500 randomly selected samples per cell type are used for this step. Genes with a Q-value > 0.05 (two-sided Wilcox test, with Benjamini-Hochberg correction for multiple hypothesis correction) are filtered out from each cell type. Of note, this filter is not necessary when discovering cell states in cell type specific profiles purified using CIBERSORTx high resolution (e.g. Tutorial 4), as CIBERSORTx incorporates its own filter for genes without evidence of expression in a given cell type. We do recommend applying it if cell type specific profiles were obtained through FACS-sorting or other deconvolution tool that does not filter for cell type specific genes.
-
Cell state discovery: EcoTyper leverages nonnegative matrix factorization (NMF) to identify transcriptionally-defined cell states from cell type specific expression profiles (step 1). Given c cell types, let
be a
cell type-specific expression matrix for cell type
consisting of
rows (the number of genes) and
columns (the number of samples). The primary objective of NMF is to factorize
into two non-negative matrices: a
matrix,
, and a
matrix,
, where
is a user-specified rank (i.e., number of clusters). The basis matrix,
, encodes a representative expression level for each gene in each cell state. The mixture coefficients matrix
, scaled to sum to 1 across cell states, encodes the representation (relative abundance) of each cell state in each sample.
EcoTyper applies NMF on the top 1000 genes with highest relative dispersion across samples. If less than 1000 genes are available, all genes are selected. If less than 50 genes are imputed for a given cell type, that cell type is not used for cell state identification. Prior to NMF, each gene is scaled to mean 0 and unit variance. To satisfy the non-negativity requirement of NMF, cell type-specific expression matrices are individually processed using posneg transformation. This function converts an input expression matrixinto two matrices, one containing only positive values and the other containing only negative values with the sign inverted. These two matrices are subsequently concatenated to produce
.
For each cell type, EcoTyper applies NMF across a range of ranks (number of cell states), by default 2-20 states. For each rank, the NMF algorithm is applied multiple times (we recommend at least 50) with different starting seeds, for robustness. -
Choosing the number of cell states: Cluster (state) number selection is an important consideration in NMF applications. We found that previous approaches that rely on minimizing error measures (e.g., RMSE, KL divergence) or optimizing information-theoretic metrics either failed to converge or were dependent on the number of genes imputed. In contrast, the cophenetic coefficient quantifies the classification stability for a given rank (i.e., the number of clusters) and ranges from 0 to 1, with 1 being maximally stable. While the rank at which the cophenetic coefficient starts decreasing is typically selected, this approach is challenging to apply in situations where the cophenetic coefficient exhibits a multi-modal shape across ranks, as we found for some cell types. Therefore, we developed a heuristic approach more suitable for such settings. In each case, the rank was automatically chosen based on the cophenetic coefficient evaluated in the range 2–20 clusters (by default). Specifically, we determined the first occurrence in the interval 2–20 for which the cophenetic coefficient dropped below 0.95 (by default), having been above this level for at least two consecutive ranks. We then selected the rank immediately adjacent to this crossing point which was closest to 0.95 (by default).
-
Extracting cell state information: The NMF output resulting from step 2 is parsed and cell state information is extracted for the downstream analyses.
-
Cell state QC filter: Although posneg transformation is required to satisfy the non-negativity constraint of NMF following standardization, it can lead to the identification of spurious cell states driven by features with more negative values than positive ones. To combat this, we devised an adaptive false positive index (AFI), a novel index defined as the ratio between the sum of weights from the W matrix corresponding to the negative and positive features. EcoTyper automatically filters the states with
.
-
Advanced cell state QC filter: When the discovery dataset is comprised of multiple tumor types, we recommend using this advanced filter. This filter identifies poor-quality cell states using a dropout score, which flags states whose marker genes exhibit anomalously low variance and high expression across the discovery cohort, generally an artefact of CIBEROSRTx HiRes. To calculate the dropout score for each marker gene (i.e., genes with maximal log2 fold change in each state relative to other states within a given cell type), EcoTyper determines the maximum fraction of samples for which the gene has the same value. It also calculates the average log2 expression of the gene across samples. It averages each quantity, scaled to unit variance across states, within each state, converts them to z-scores, and removes states with a mean Z >1.96 (P < 0.05).
-
Ecotype (cellular community) discovery: Ecotypes or cellular communities are derived by identifying patterns of co-occurrence of cell states across samples. First, EcoTyper leverages the Jaccard index to quantify the degree of overlap between each pair of cell states across samples in the discovery cohort. Toward this end, it discretizes each cell state
into a binary vector
of length
, where
= the number of samples in the discovery cohort. Collectively, these vectors comprise binary matrix
, with same number of rows as cell states across cell types and
columns (samples). Given sample
, if state
is the most abundant state among all states in cell type
, EcoTyper sets
to 1; otherwise
. It then computes all pairwise Jaccard indices on the rows (states) in matrix
, yielding matrix
. Using the hypergeometric test, it evaluates the null hypothesis that any given pair of cell states
and
have no overlap. In cases where the hypergeometric p-value is >0.01, the Jaccard index for
is set to 0 (i.e., no overlap). To identify communities while accommodating outliers, the updated Jaccard matrix
is hierarchically clustered using average linkage with Euclidean distance (hclust in the R stats package). The optimal number of clusters is then determined via silhouette width maximization. Clusters with less than 3 cell states are eliminated from further analysis.
In order for EcoTyper to perform cell states and ecotypes discovery, the following resources need to be available:
-
user-provided cell-type specific expression matrices, on which the discovery will be performed (a discovery cohort). For this tutorial, we will use the example data in
example_data/Tutorial_6/PresortedDiscovery. -
optionally, a sample annotation file, such as the one provided in
example_data/Tutorial_6/PresortedDiscovery_annotation.txt, can be supplied to EcoTyper. The information in this file can be used for heatmap plotting purposes, and also to instruct EcoTyper to find cell states/ecotypes common across different biological batches (e.g. tumor types), as detailed below.
The script that does cell type and ecotype discovery is:
Rscript EcoTyper_discovery_presorted.R -h## usage: EcoTyper_discovery_presorted.R [-c <PATH>] [-h]
##
## Arguments:
## -c <PATH>, --config <PATH>
## Path to the config files [required].
## -h, --help Print help message.
This script takes as input file a configuration file in
YAML format. The configuration file for this
tutorial is available in config_discovery_presorted.yml:
default :
Input :
Discovery dataset name : "PresortedDiscovery"
Expression matrices : "example_data/Tutorial_6/PresortedDiscovery"
Annotation file : "example_data/Tutorial_6/PresortedDiscovery_annotation.txt"
Annotation file column to scale by : "Histology"
Annotation file column(s) to plot : ["Histology", "Tissue"]
Output :
Output folder : "PresortedDiscoveryOutput"
Pipeline settings :
#Pipeline steps:
# step 1 (extract cell type specific genes)
# step 2 (cell state discovery)
# step 3 (choosing the number of cell states)
# step 4 (extracting cell state information)
# step 5 (cell state QC filter)
# step 6 (advanced cell state QC filter)
# step 7 (ecotype discovery)
Pipeline steps to skip : [6,]
Filter non cell type specific genes : False
Number of threads : 10
Number of NMF restarts : 5
Maximum number of states per cell type : 20
Cophenetic coefficient cutoff : 0.95
The configuration file has three sections, Input, Output and Pipeline settings. We next will describe the expected content in each of these three sections, and instruct the user how to set the appropriate settings in their applications.
The Input section contains settings regarding the input data.
Discovery dataset name is the identifier used by EcoTyper to
internally save and retrieve the information about the cell
states/ecotypes defined on this discovery dataset. It is also the name
to be provided to the -d/–discovery argument of scripts
EcoTyper_recovery_scRNA.R and EcoTyper_recovery_bulk.R, when
performing cell state/ecotypes recovery. Any value that contains
alphanumeric characters and ’_’ is accepted for this field.
Discovery dataset name : "PresortedDiscovery"Expression matrices : "example_data/Tutorial_6/PresortedDiscovery"Expression matrices field should contain the path to directory with a tab-delimited file containing cell type specific expression data for each cell type. Each file should have genes as rows and samples as columns, should be in the TPM or FPKM space for bulk RNA-seq and non-logarithmic (exponential) space for microarrays. They should have gene symbols on the first column and gene counts for each sample on the next columns. Column (sample) names should be unique within each file. The same sample ids (column names) should be present in each cell type specific matrix. Also, we recommend that the column names do not contain special characters that are modified by the R function make.names, e.g. having digits at the beginning of the name or containing characters such as space, tab or -:
The expected format for each expression matrix is:
data = read.delim("example_data/Tutorial_6/PresortedDiscovery/Fibroblasts.txt", nrow = 5)
dim(data)## [1] 5 251
head(data[,1:5])## GeneSymbol TCGA.37.A5EN.01A.21R.A26W.07 TCGA.37.4133.01A.01R.1100.07
## 1 A1BG 29.356911 29.220771
## 2 AAR2 47.746044 47.746617
## 3 ABCA6 5.803932 5.413472
## 4 ABCB7 23.299299 25.486127
## 5 ABI2 37.677476 32.007233
## TCGA.77.7465.01A.11R.2045.07 TCGA.34.5240.01A.01R.1443.07
## 1 29.228389 28.835613
## 2 47.828573 47.679150
## 3 6.259786 6.092511
## 4 43.447296 41.077193
## 5 31.775814 31.421975
Annotation file : "example_data/Tutorial_6/PresortedDiscovery_annotation.txt"A path to an annotation file can be provided in the field Annotation file. If provided, this file should contain a column called ID with the same names as the columns of the expression matrices, and any number of additional columns. The additional columns can be used for defining sample batches (see Section Annotation file column to scale by below) and for plotting color bars in the heatmaps output (see Section Annotation file column(s) to plot below). If not provided, this field needs to be set to “NULL”. For the current example, the annotation file has the following format:
annotation = read.delim("example_data/Tutorial_6/PresortedDiscovery_annotation.txt", nrow = 5)
dim(annotation)## [1] 5 6
head(annotation)## ID Tissue Histology Type OS_Time
## 1 TCGA.37.A5EN.01A.21R.A26W.07 Tumor LUSC Primary Solid Tumor 660
## 2 TCGA.37.4133.01A.01R.1100.07 Tumor LUSC Primary Solid Tumor 238
## 3 TCGA.77.7465.01A.11R.2045.07 Tumor LUSC Primary Solid Tumor 990
## 4 TCGA.34.5240.01A.01R.1443.07 Tumor LUSC Primary Solid Tumor 1541
## 5 TCGA.05.4249.01A.01R.1107.07 Tumor LUAD Primary Solid Tumor 1523
## OS_Status
## 1 0
## 2 0
## 3 0
## 4 0
## 5 0
Annotation file column to scale by : "Histology"In order to discover pan-carcinoma cell states and ecotypes in the EcoType carcinoma paper, we standardize genes to mean 0 and unit 1 within each tumor type (histology). Field Annotation file column to scale by allows users to specify a column name in the annotation file, by which the samples will be grouped when performing standardization. The example discovery dataset used in this tutorial has samples from lung adenocarcinoma and lung squamous cell carcinoma. Therefore, for this tutorial we will use the Histology column to perform standardization.
However, this is an analytical choice, depending on the purpose of the analysis. If the users are interested in defining cell states and ecotypes regardless of tumor type-specificity, this argument can be set to “NULL”. In this case, the standardization will be applied across all samples in the discovery cohort. The same will happen if the annotation file is not provided.
Annotation file column(s) to plot : ["Histology", "Tissue"]Annotation file column(s) to plot field specifies which columns in the annotation file will be used as color bar in the output heatmaps, in addition to the cell state label or ecotype label column, plotted by default.
The Output section contains a single field, Output folder, which specifies the path where the final output will be saved. This folder will be created if it does not exist.
Output folder : "PresortedDiscoveryOutput"The last section, Pipeline settings, contains settings controlling how EcoTyper is run.
Pipeline steps:
# step 1 (extract cell type specific genes)
# step 2 (cell state discovery)
# step 3 (choosing the number of cell states)
# step 4 (extracting cell state information)
# step 5 (cell state QC filter)
# step 6 (advanced cell state QC filter)
# step 7 (ecotype discovery)
Pipeline steps to skip : [6,] #by default, step 6 is skippedThe Pipeline steps to skip option allows user to skip some of the steps outlined in section Overview of the EcoTyper workflow for discovering cell states. Please note that this option is only intended for cases when the pipeline had already been run once, and small adjustments are made to the parameters. For example, if the Cophenetic coefficient cutoff used in step 3 needs adjusting, the user might want to skip steps 1-2 and re-run from step 3 onwards.
Filter non cell type specific genes : FalseFlag indicated whether to apply the filter for cell type specific genes in step 1, outlined in section Overview of the EcoTyper workflow for discovering cell states. This filter is not necessary when discovering cell states in cell type specific profiles purified using CIBERSORTx high resolution (e.g. Tutorial 4), as CIBERSORTx incorporates its own filter for genes without evidence of expression in a given cell type. We do recommend applying it if cell type specific profiles were obtained through FACS-sorting or other deconvolution tool that does not filter for cell type specific genes.
We set it to False in this tutorial, as the input matrices were obtained using CIBERSORTx.
Number of threads : 10The number of threads EcoTyper will be run on.
Number of NMF restarts : 5The NMF approach used by EcoTyper (Brunet et al.), can give slightly different results, depending on the random initialization of the algorithm. To obtain a stable solution, NMF is generally run multiple times with different seeds, and the solution that best explains the discovery data is chosen. Additionally, the variation of NMF solutions across restarts with different seeds is quantified using Cophenetic coefficients and used in step 4 of EcoTyper for selecting the number of states. The parameter Number of NMF restarts specifies how many restarts with different seed should EcoTyper perform for each rank selection, in each cell type. Since this is a very time consuming process, in this example we only use 5. However, for publication-quality results, we recommend at least 50 restarts.
Maximum number of states per cell type : 20Maximum number of states per cell type specifies the upper end of the range for the number of states possible for each cell type. The lower end is 2.
Cophenetic coefficient cutoff : 0.95This field indicates the Cophenetic coefficient cutoff, in the range [0, 1], used for automatically determining the number of states in step 4. Lower values generally lead to more clusters being identified.
After editing the configuration file (config_discovery_presorted.yml),
the de novo discovery cell states and ecotypes from presorted
expression profiles can be run as is illustrated below. Please note that
this script might take up to two hours to run on 10 threads. Also,
although EcoTyper can be run on the example data from this tutorial
using a typical laptop (16GB memory), it might not be the case for
larger datasets. We recommend that cell type and ecotype discovery is
generally run on a server with >32GB of RAM.
Rscript EcoTyper_discovery_presorted.R -c config_discovery_presorted.ymlEcoTyper generates for each cell type the following outputs:
-
Plots displaying the Cophenetic coefficient calculated in step 4. The horizontal dotted line indicates the Cophenetic coefficient cutoff provided in the configuration file Cophenetic coefficient cutoff field. The vertical dotted red line indicates the number of states automatically selected based on the Cophenetic coefficient cutoff provided. We recommend that users inspect this file to make sure that the automatic selection provides sensible results. If the user wants to adjust the Cophenetic coefficient cutoff after inspecting this plot, they can rerun the discovery procedure skipping steps 1-3. Please note that:
- These plots indicate the number of states obtained before applying the filters for low-quality states in steps 6 and 7. Therefore, the final results will probably contain fewer states.
- The plots below might look slightly different when generated with R versions other than R/3.6.0. This is because some EcoTyper steps, including NMF algorithm initialization, depend on random number generation. Althoguh EcoTyper sets random seeds before each such step, different R version output different random numbers for the same seed. To mitigate this issue, we recommend at least 50 NMF restarts when running EcoTyper.
knitr::include_graphics("PresortedDiscoveryOutput/rank_plot.png")-
For each cell type, the following outputs, exemplified here for endothelial cells, are produced:
- Abundances of cell states remaining after the QC filters in steps 6 and 7 (if run), across samples in the discovery dataset:
data = read.delim("PresortedDiscoveryOutput/Endothelial.cells/state_abundances.txt")
dim(data)## [1] 4 250
head(data[,1:5])## TCGA.37.A5EN.01A.21R.A26W.07 TCGA.37.4133.01A.01R.1100.07
## S01 4.657038e-15 3.396931e-15
## S02 4.313475e-01 3.396931e-15
## S03 4.657038e-15 3.396931e-15
## S04 5.532795e-02 3.396931e-15
## TCGA.77.7465.01A.11R.2045.07 TCGA.34.5240.01A.01R.1443.07
## S01 4.011227e-15 3.955821e-15
## S02 2.750005e-01 8.143772e-02
## S03 4.011227e-15 3.955821e-15
## S04 4.011227e-15 1.748715e-03
## TCGA.05.4249.01A.01R.1107.07
## S01 4.256051e-15
## S02 4.256051e-15
## S03 1.137231e-01
## S04 8.575277e-01
- Assignment of samples in the discovery dataset to the cell state with the highest abundance. Only samples assigned to the cell states remaining after the QC filters in steps 6 and 7 (if run) are included. The remaining ones are considered unassigned and removed from this table:
data = read.delim("PresortedDiscoveryOutput/Endothelial.cells/state_assignment.txt")
dim(data)## [1] 131 3
head(data)## ID State InitialState
## 31 TCGA.55.6983.11A.01R.1949.07 S01 IS02
## 32 TCGA.44.6776.11A.01R.1858.07 S01 IS02
## 33 TCGA.77.7335.11A.01R.2045.07 S01 IS02
## 34 TCGA.38.A44F.01A.11R.A24H.07 S01 IS02
## 35 TCGA.77.7138.11A.01R.2045.07 S01 IS02
## 36 TCGA.44.6778.11A.01R.1858.07 S01 IS02
- A heatmap illustrating the expression of genes used for cell state discovery, that have the highest fold-change in one of the cell states remaining after the QC filters in steps 6 and 7 (if run). In the current example, the heatmap includes in the top color bar two rows corresponding to Tissue and Histology, that have been provided in configuration file field Annotation file column(s) to plot, in addition to cell state labels always plotted:
knitr::include_graphics("PresortedDiscoveryOutput/Endothelial.cells/state_assignment_heatmap.png")The ecotype output files include:
- The cell state composition of each ecotype (the set of cell states making up each ecotype):
ecotypes = read.delim("PresortedDiscoveryOutput/Ecotypes/ecotypes.txt")
head(ecotypes[,c("CellType", "State", "Ecotype")])## CellType State Ecotype
## 1 B.cells S01 E1
## 2 Endothelial.cells S02 E1
## 3 Epithelial.cells S01 E1
## 4 Fibroblasts S07 E1
## 5 B.cells S03 E2
## 6 CD4.T.cells S02 E2
- The number of initial clusters obtained by clustering the Jaccard index matrix, selected using the average silhouette:
knitr::include_graphics("PresortedDiscoveryOutput/Ecotypes/nclusters_jaccard.png")- A heatmap of the Jaccard index matrix, after filtering ecotypes with less than 3 cell states:
knitr::include_graphics("PresortedDiscoveryOutput/Ecotypes/jaccard_matrix.png")- The abundance of each ecotype in each sample in the discovery dataset:
abundances = read.delim("PresortedDiscoveryOutput/Ecotypes/ecotype_abundance.txt")
dim(abundances)## [1] 7 250
head(abundances[,1:5])## TCGA.37.A5EN.01A.21R.A26W.07 TCGA.37.4133.01A.01R.1100.07
## E1 0.794531073 5.587512e-02
## E2 0.013996811 7.309504e-02
## E3 0.069214903 6.880715e-03
## E4 0.003306485 6.049462e-03
## E5 0.025837936 1.759492e-14
## E6 0.093112792 8.580997e-01
## TCGA.77.7465.01A.11R.2045.07 TCGA.34.5240.01A.01R.1443.07
## E1 4.508978e-01 9.485239e-01
## E2 3.120952e-01 2.957054e-03
## E3 7.323729e-15 1.835542e-03
## E4 8.909708e-15 1.377273e-02
## E5 1.645593e-01 7.296796e-15
## E6 7.244779e-02 3.291079e-02
## TCGA.05.4249.01A.01R.1107.07
## E1 4.493338e-09
## E2 1.180982e-01
## E3 5.358678e-01
## E4 1.804165e-01
## E5 8.538532e-02
## E6 1.303634e-15
- The assignment of samples in the discovery dataset to ecotypes. The samples not assigned to any ecotype are filtered out from this file:
assignments = read.delim("PresortedDiscoveryOutput/Ecotypes/ecotype_assignment.txt")
dim(assignments)## [1] 190 11
head(assignments[,1:5])## ID MaxEcotype
## TCGA.22.4613.01A.01R.1443.07 TCGA.22.4613.01A.01R.1443.07 E1
## TCGA.22.5471.01A.01R.1635.07 TCGA.22.5471.01A.01R.1635.07 E1
## TCGA.22.5473.01A.01R.1635.07 TCGA.22.5473.01A.01R.1635.07 E1
## TCGA.34.5240.01A.01R.1443.07 TCGA.34.5240.01A.01R.1443.07 E1
## TCGA.34.8456.01A.21R.2326.07 TCGA.34.8456.01A.21R.2326.07 E1
## TCGA.37.A5EM.01A.21R.A27Q.07 TCGA.37.A5EM.01A.21R.A27Q.07 E1
## AssignmentP AssignmentQ AssignedToEcotypeStates
## TCGA.22.4613.01A.01R.1443.07 0.006237509 0.02267708 TRUE
## TCGA.22.5471.01A.01R.1635.07 0.011589892 0.03408792 TRUE
## TCGA.22.5473.01A.01R.1635.07 0.173391227 0.21892832 TRUE
## TCGA.34.5240.01A.01R.1443.07 0.073719200 0.12710207 TRUE
## TCGA.34.8456.01A.21R.2326.07 0.002977868 0.01404655 TRUE
## TCGA.37.A5EM.01A.21R.A27Q.07 0.027084554 0.06573921 TRUE
- A heatmap of cell state fractions across the samples assigned to ecotypes:
knitr::include_graphics("PresortedDiscoveryOutput/Ecotypes/heatmap_assigned_samples_viridis.png")Question: How do I run EcoTyper on a high-performance cluster,
rather than a single server?
Answer: EcoTyper can be modified
to run on a high-performance cluster, by overriding the
pipeline\lib\multithreading.R library. Currently the library provides
two functions, PushToJobQueue which adds a command line call to the
job queue, and RunJobQueue which waits for all the jobs in the queue
to finish. The default implementation of these functions uses R function
mclapply to perform computations on multiple threads:
job_queue = c()
PushToJobQueue <- function(cmd){
job_queue <<- c(job_queue, cmd)
}
RunJobQueue <- function()
{
if(length(job_queue) == 0)
{
return(NULL)
}
res = mclapply(job_queue, FUN = system, mc.cores = n_threads)
job_queue <<- c()
errors = sum(unlist(res))
if(errors > 0)
{
stop("EcoTyper failed. Please check the error message above!")
}
}Users can re-write these two functions according to the requirements of their cluster infrastructure. A primitive example of how this can be achieved on a high performance cluster built on the SLURM infrastructure is:
PushToJobQueue <- function(cmd){
system(paste0("Rscript run_job.R ", cmd))
}
RunJobQueue <- function()
{
name_substr = discovery
while(job_is_running(name_substr))
{
print("Sleeping 60s...")
Sys.sleep(60)
}
}
job_is_running <- function(name_substr)
{
while(T)
{
possibleError <- tryCatch({
out = system("squeue -o '%.18i\t%.9P\t%j\t%.8u\t%.8T\t%.10M\t%.9l\t%.6D\t%R'", intern = T)
con <- textConnection(out)
data <- read.delim(con)
if(ncol(data) < 9)
{
Sys.sleep(30)
next
}
response = sum(grepl(name_substr, data[,3])) > 0
return(response)
}, error = function(e){})
if(inherits(possibleError, "error"))
{
next
}
}
}Where run_job.R is a script that takes as input a command line and
submits the job to cluster:
template = '#!/bin/bash
#SBATCH --job-name=<TMP>
#SBATCH --begin=now
#SBATCH --time=3:00:00
#SBATCH --mem=30G
#SBATCH -p normal
#SBATCH -c 1
#SBATCH --error=../jobs/<R_SCRIPT_NAME>/<TMP>.err
#SBATCH --output=../jobs/<R_SCRIPT_NAME>/<TMP>.out
<R_SCRIPT> <ARGUMENTS>
'
args <- commandArgs(T)
script_name = args[1]
arguments = args[-1]
output = file.path("../jobs", basename(script_name))
dir.create(output, recursive = T, showWarning = F)
arguments_s = ifelse(grepl("/", arguments, fixed = T), basename(arguments), arguments)
tmp = paste(arguments_s, collapse = "_")
job = gsub("<R_SCRIPT>", script_name, template, fixed = T)
job = gsub("<R_SCRIPT_NAME>", basename(script_name), job, fixed = T)
job = gsub("<ARGUMENTS>", paste0(arguments, collapse = " "), job, fixed = T)
job = gsub("<TMP>", tmp, job, fixed = T)
output_path <- file.path(output, paste0(tmp, ".sh"))
write(job, output_path)
system(paste0("sbatch ", output_path))## Warning in system(paste0("sbatch ", output_path)): error in running command