![]() CoNekT, the Coexpression Network Toolkit,
documentation and code.
CoNekT, the Coexpression Network Toolkit,
documentation and code.
If you use CoNekT in your research please cite: CoNekT: an open-source framework for comparative genomic and transcriptomic network analyses. Proost et al. 2018. ( https://doi.org/10.1093/nar/gky336 )
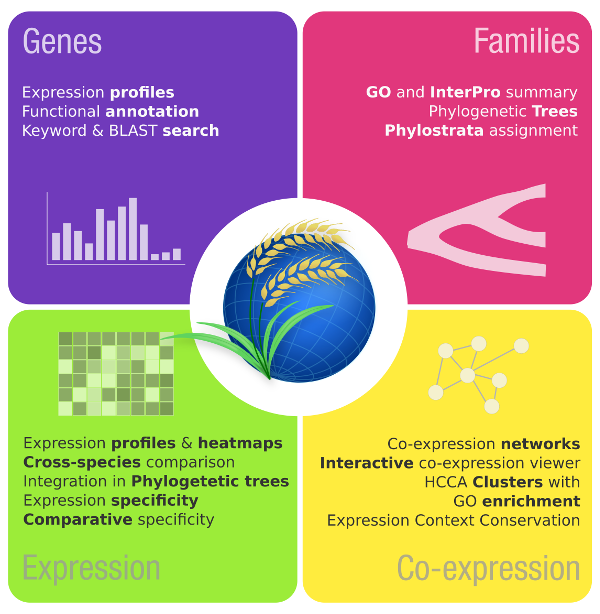
CoNekT is an online platform that allows users to browse expression profiles, study expression specificty, co-expression networks and more across different species.
Here you can find tutorials how to use the public version along with the source-code and build instructions that allow you to host your own instance with other species or in-house data.
v1.1
- General tree import included
- Updated dependencies that had known security vulnerabilities and adjusted code where necessary
- Various small tweaks
v1.0
- Initial release
- The Basics
- Expression profiles, heatmaps and specificity
- Gene Families and Phylogenetic trees
- Comparing specificity
- Coexpression Networks and Clusters
Anything missing ? Further questions ? Don't hesitate to contact us.
Below are the instructions to set up and host your own CoNekT instance.
- Linux Installation instructions
- Windows Installation instructions
- Run website
- Build CoNekT: Upload your data to the platform using the Admin interface
Optional steps (recommended for production environments)
CoNekT is based upon many open-source projects, check the full list here.
Issues are the preferred way to report bugs and errors, start a discussion for suggestions and feedback.