SMILE package implements Linear Genetic Programming (LGP) algorithm in python, with a scikit-learn style API. It is mainly used in data mining and finding feature interactions. Note it currently only support binary classification data.
Quick tutorial on SMILE framework: https://youtu.be/7sPdUTrNIZs
This package is published on pypi. Install using pip.
pip install lgpSample metabolomic data on AD can be found in dataset folder or directly downloaded from the website.
This algorithm is computationally expensive, and it needs to run approximately 1000 times in parallel to produce enough data to analyze. it needs to run in computer clusters like compute canada.
Create a running python file (Run.py) in the same directory as lgp folder, Sample Run.py and classifier usages are shown below:
from linear_genetic_programming.lgp_classifier import LGPClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn import metrics
# preprocess your data, get data matrix X, label y and names
# X, y are in scikit-learn style
X, y, names
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=1)
# set own parameter here
lgp = LGPClassifier(numberOfInput = X_train.shape[1], numberOfVariable = 200, populationSize = 20,
fitnessThreshold = 1.0, max_prog_ini_length = 40, min_prog_ini_length = 10,
maxGeneration = 2, tournamentSize = 4, showGenerationStat=False,
isRandomSampling=True, maxProgLength = 500)
lgp.fit(X_train, y_train)
y_pred = lgp.predict(X_test)
y_prob = lgp.predict_proba(X_test)[:, 0]
lgp.testingAccuracy = accuracy_score(y_pred, y_test)
# calculate F1, AUC scores
f1_scores = metrics.f1_score(y_test, y_pred, pos_label=0)
fpr, tpr, thresholds = metrics.roc_curve(y_test, y_prob, pos_label=0)
auc_scores = metrics.auc(fpr, tpr)
# store F1, AUC in validationScores
lgp.validationScores = {'f1':f1_scores, 'auc':auc_scores}
# the result can be saved by calling save_model(). It will produce a pickle file.
# save_model() use pickle for object serialization
lgp.save_model()Then use bash file to set running parameters and submit jobs. This might be different in different supercomputers. Here is an example Bash running script in Compute Canada:
#!/bin/bash
#SBATCH --time=10:00:00
#SBATCH --array=1-1000
#SBATCH --mem=500M
#SBATCH --job-name="lgp"
python Run.py.pklfile is produce (usingsave_model()method) when running the algorithm.csvis the original dataset file. Make sure you named the class column 'category' and put feature names as column names. The dataset is read using the following pandas code.
File error checking: Download error checking file input_file_error_checking.py.
Put your prepared files in the same directory and run input_file_error_checking.py.
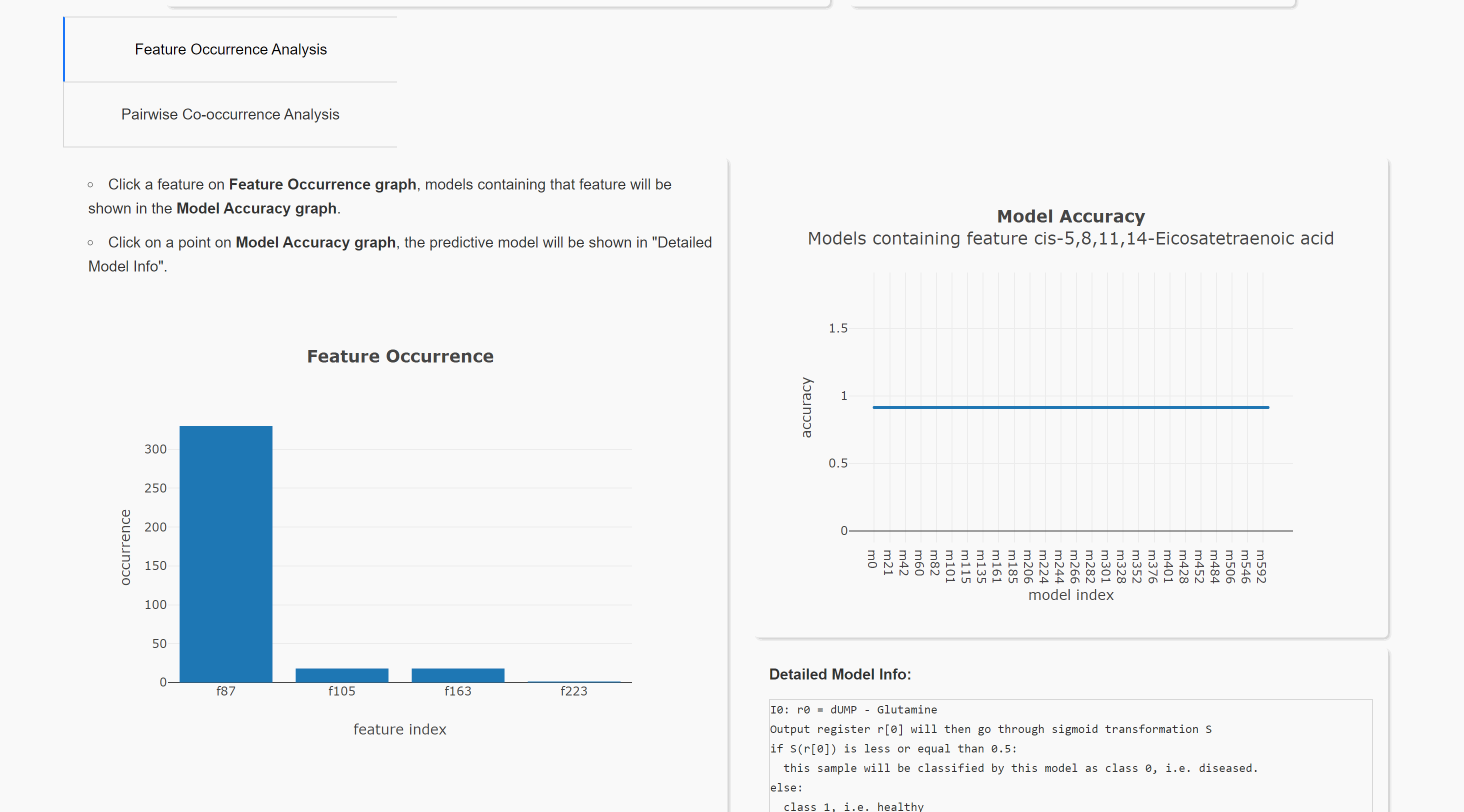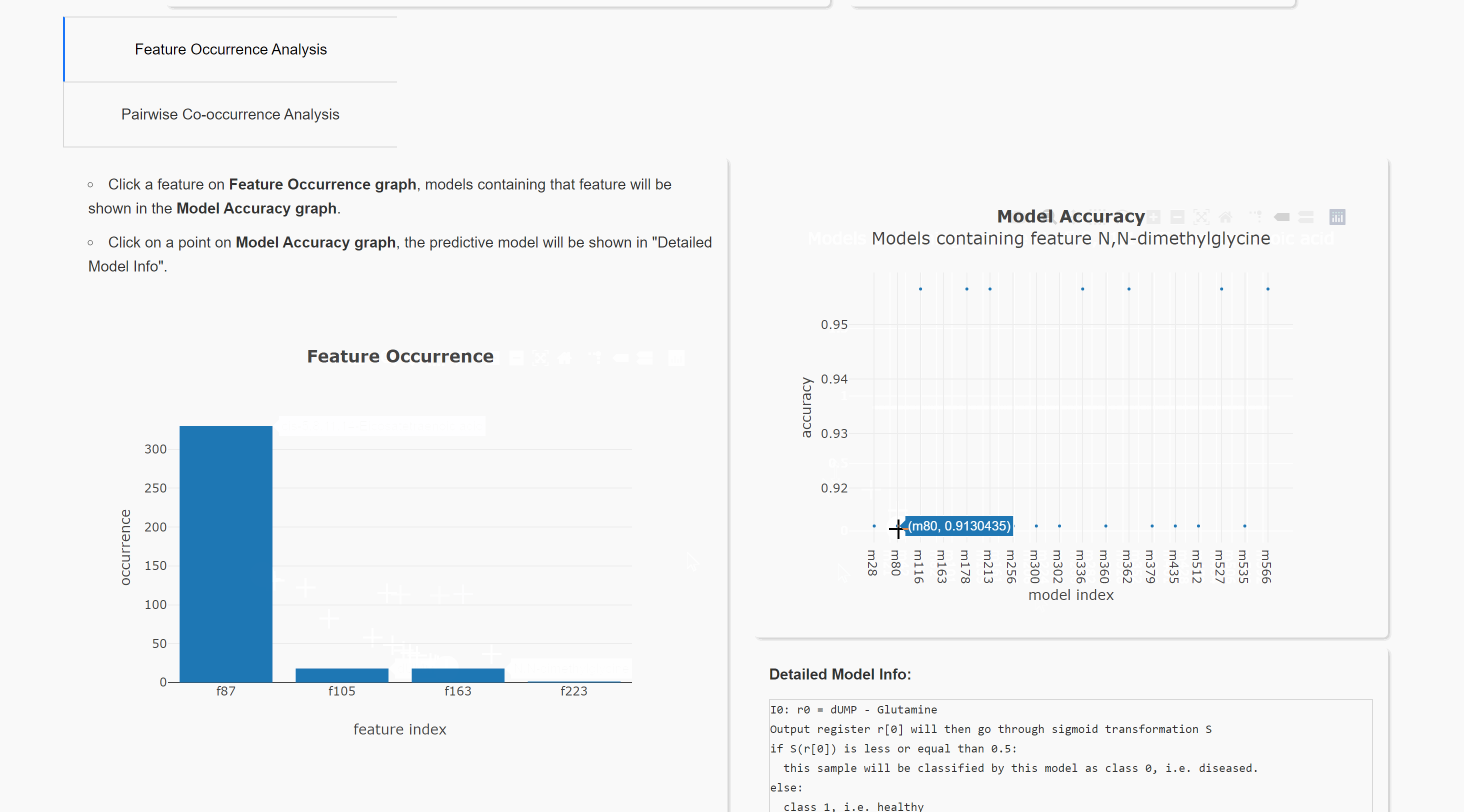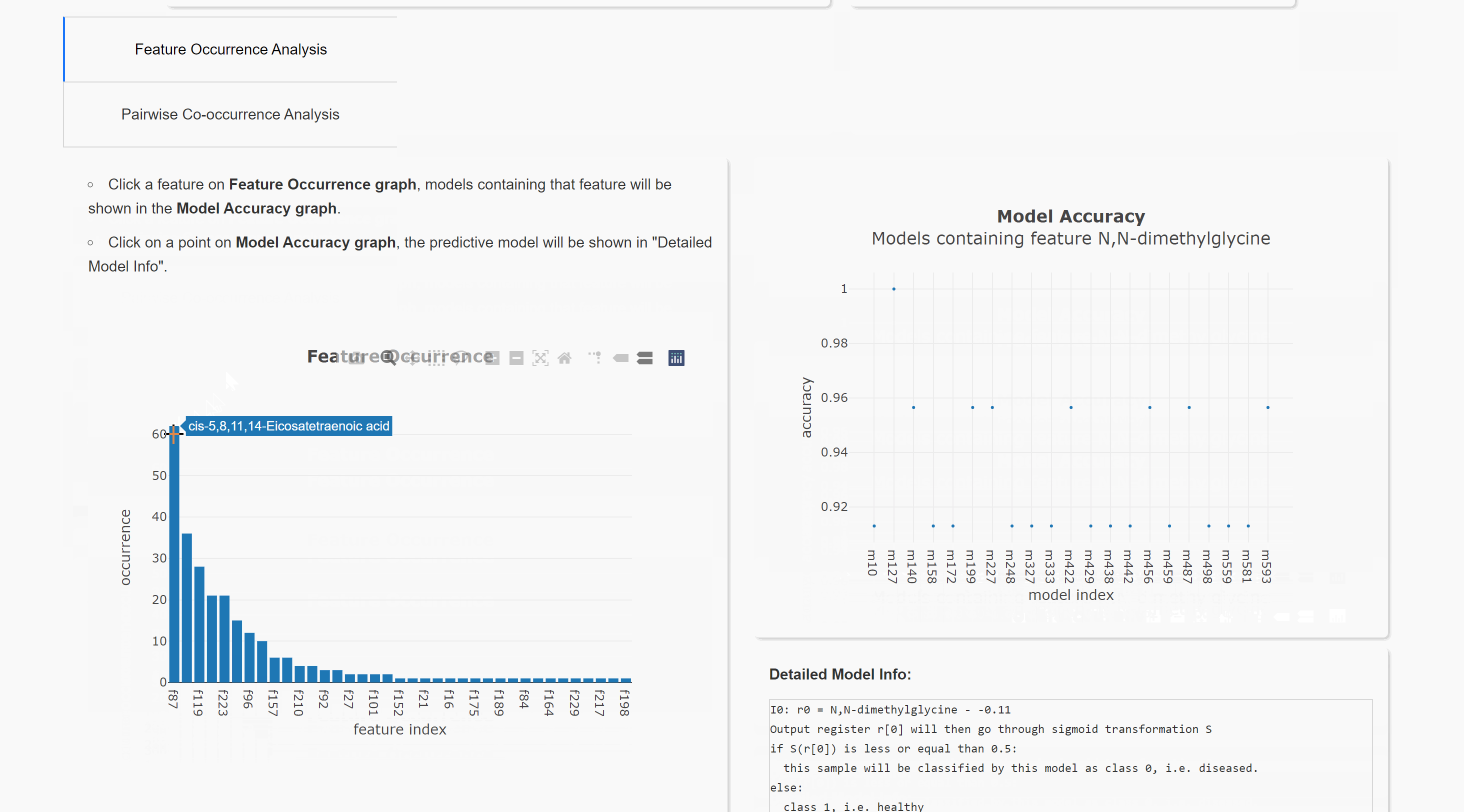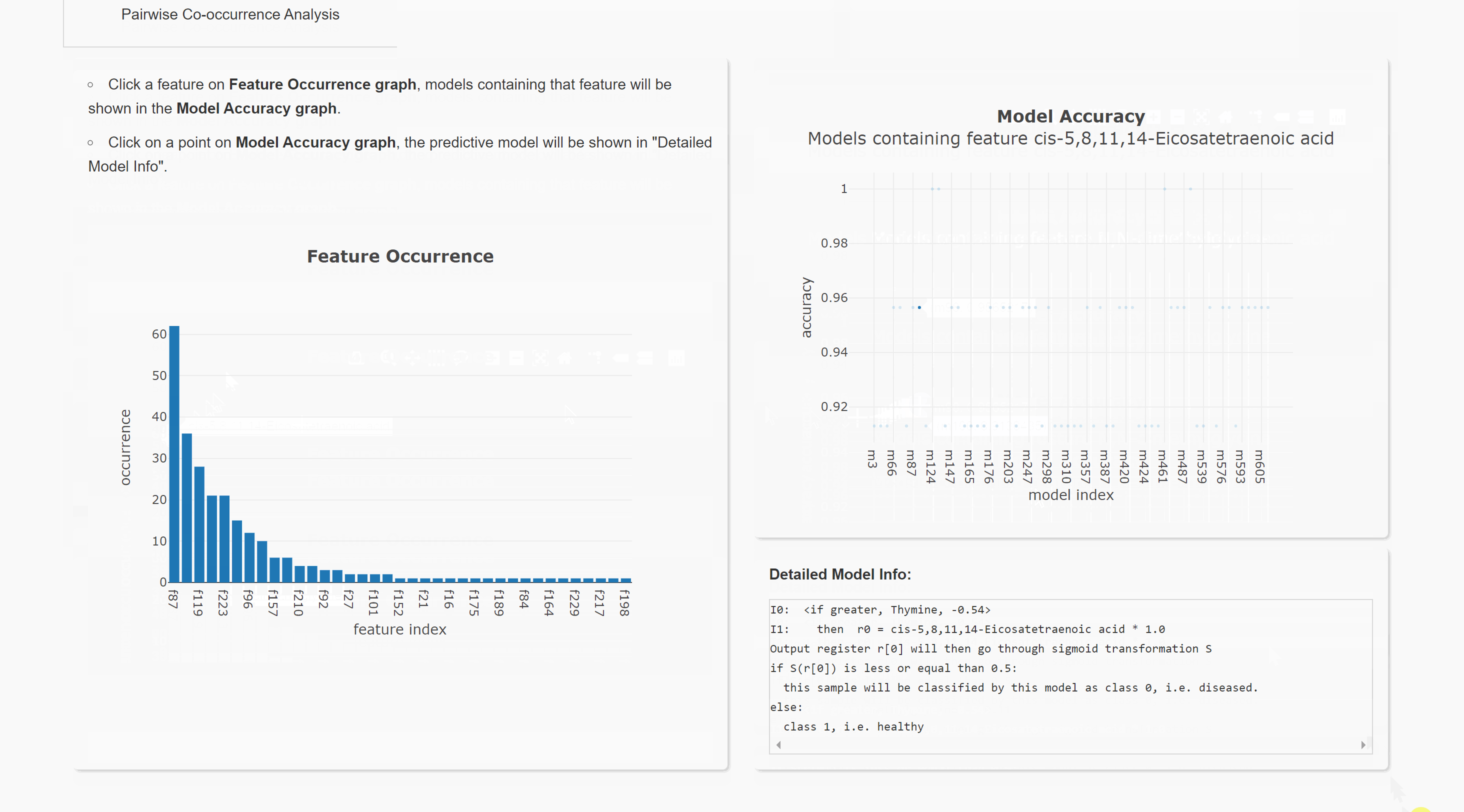
Upload result files to result visualization website (herokuapp hosting). or result visualization website (Queen's CS hosting). This will help you visualize the result. Note the herokuapp web server uses ephemeral filesystem, that means all files are lost when you restart the web.
You can also run the visualization locally. Download website source code. After installing all requirements (listed in requirements.txt), you can run this website in your local browser.
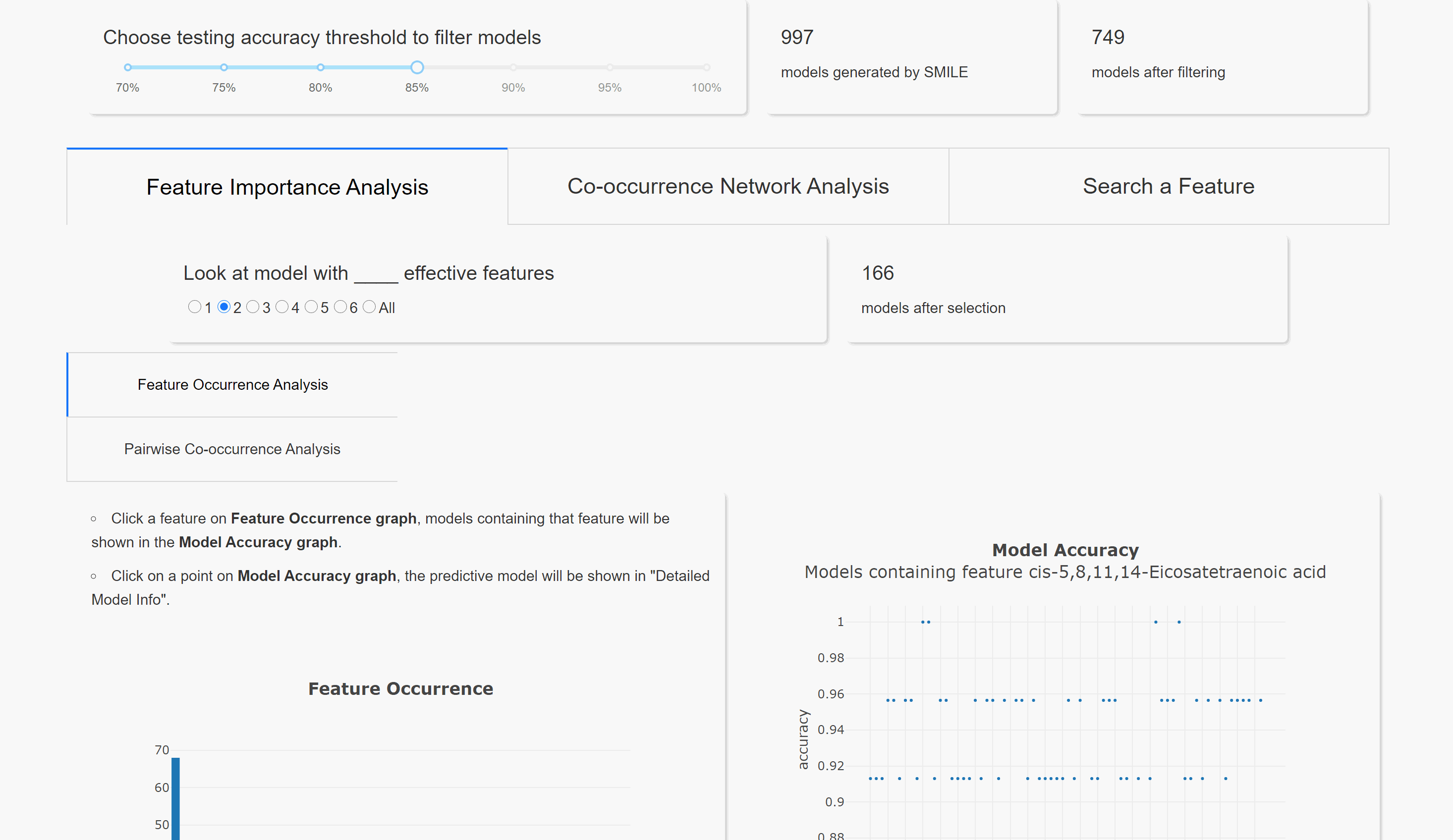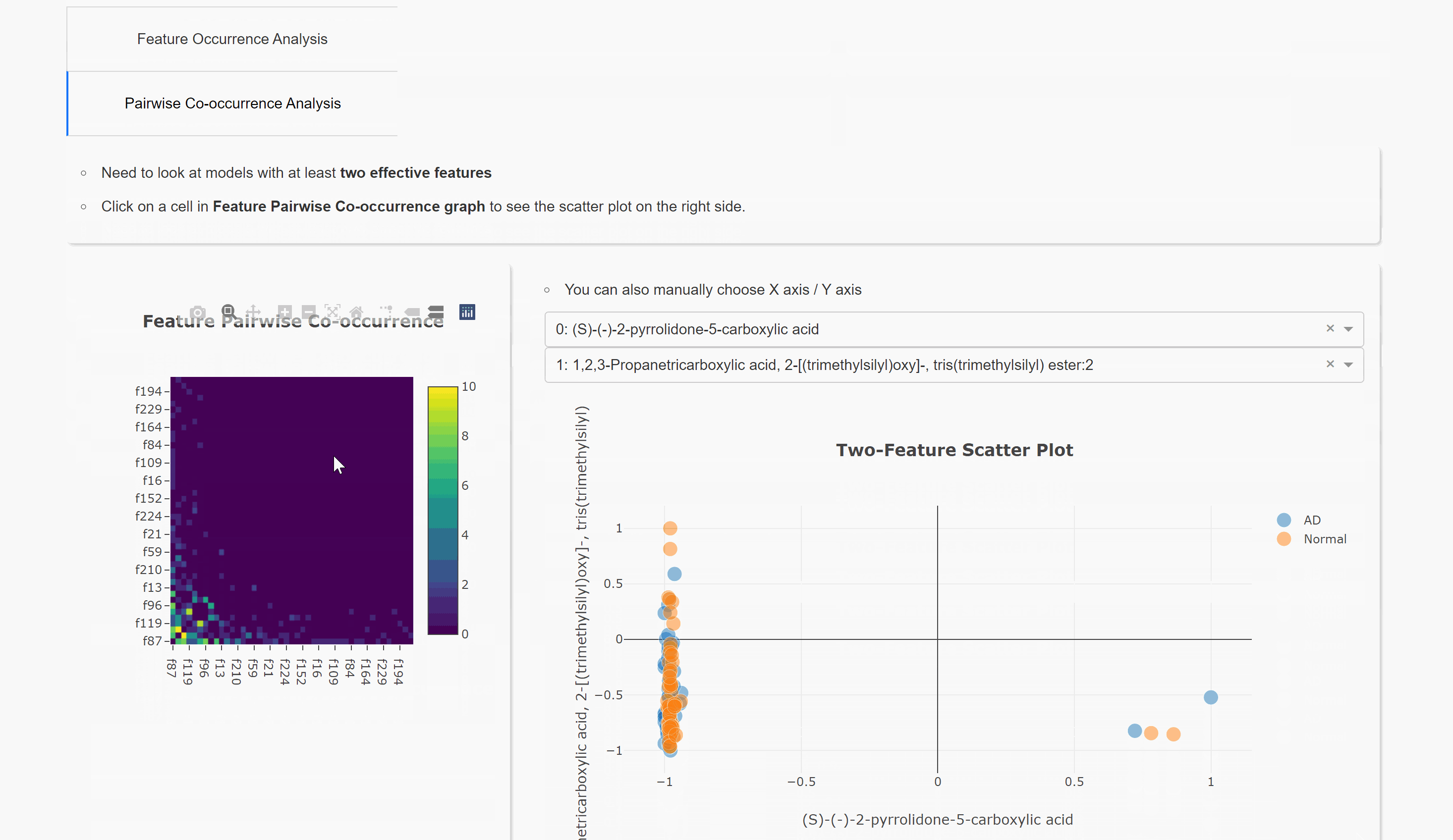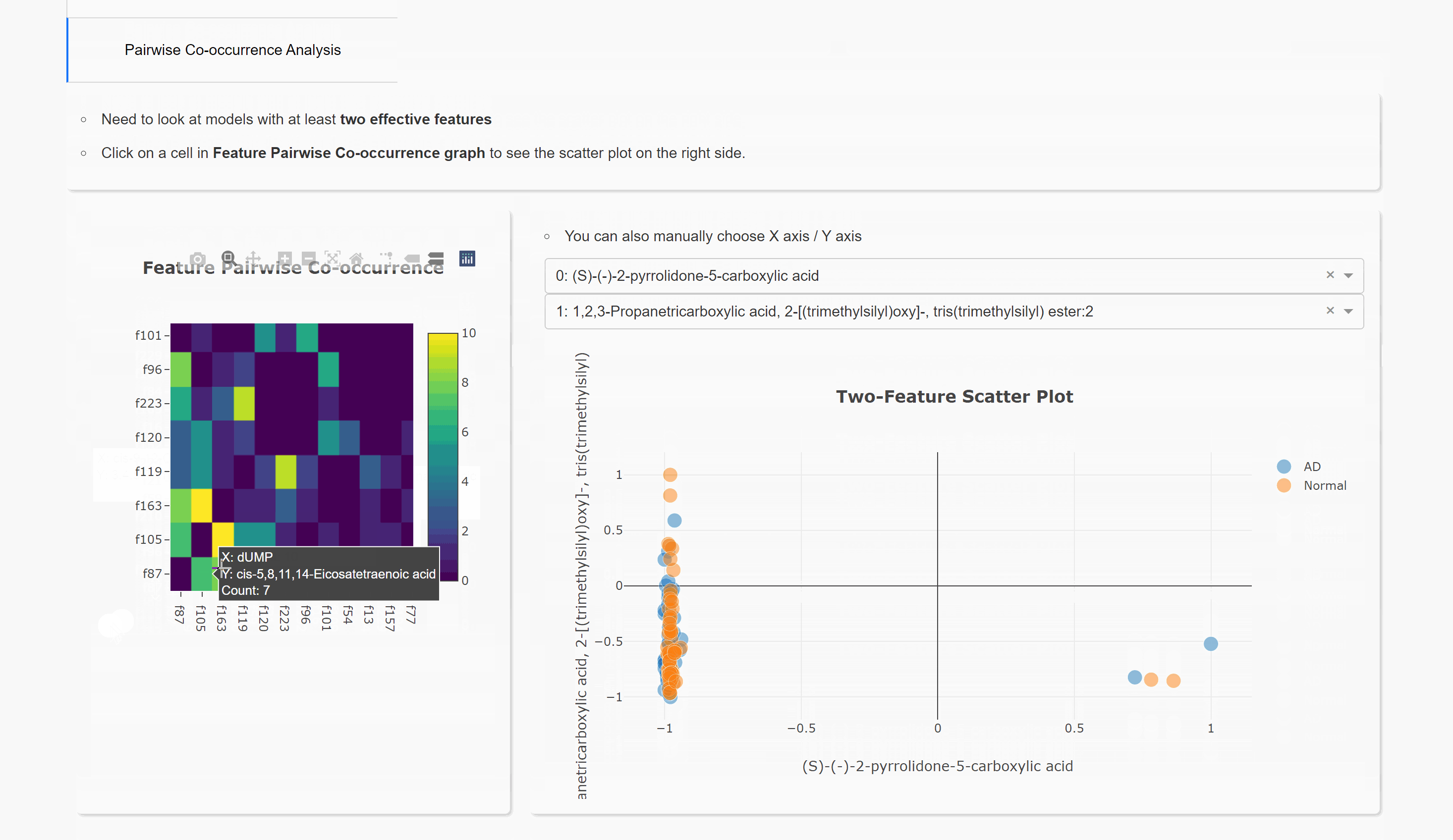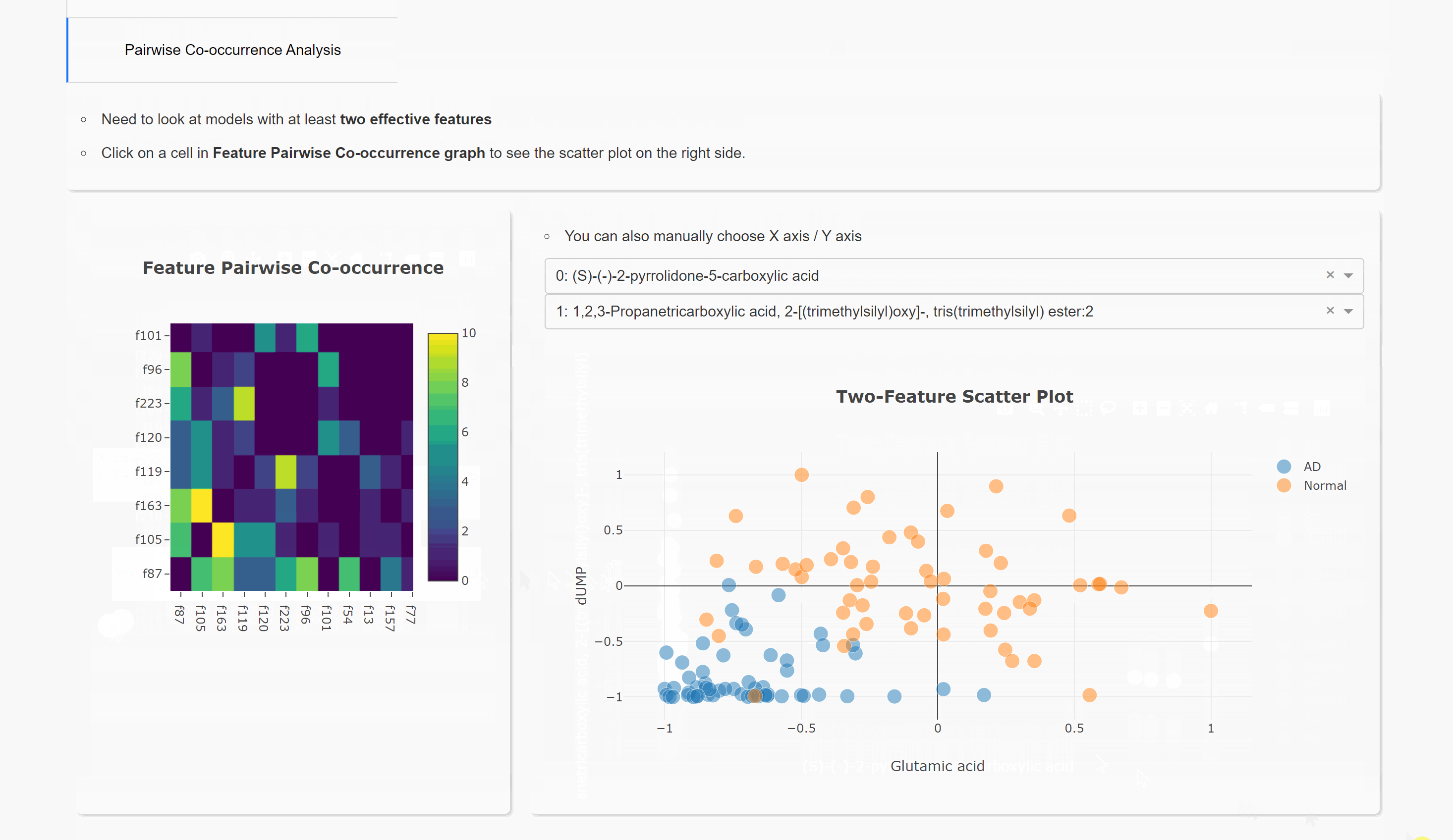
Feature Occurrence Analysis
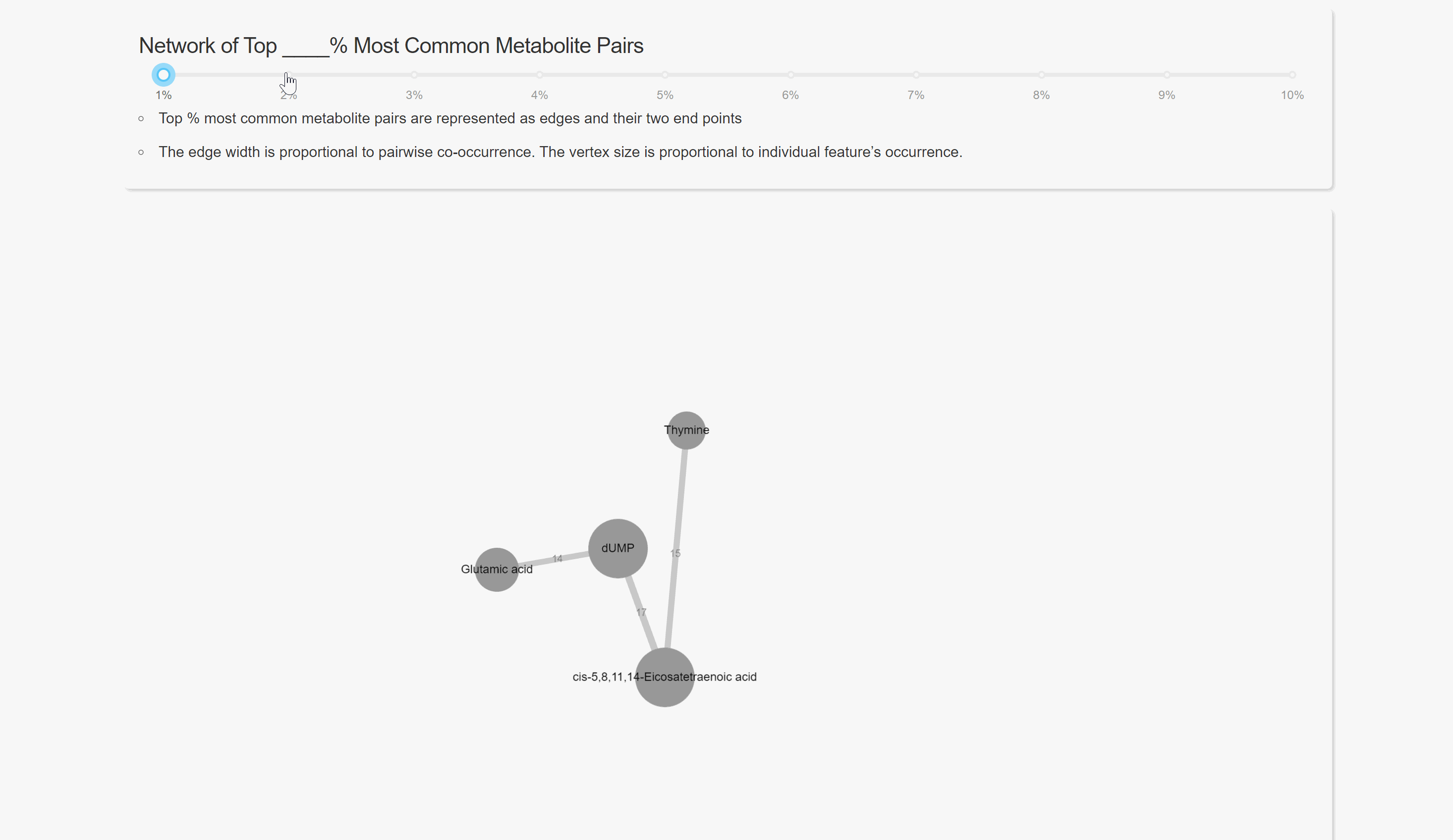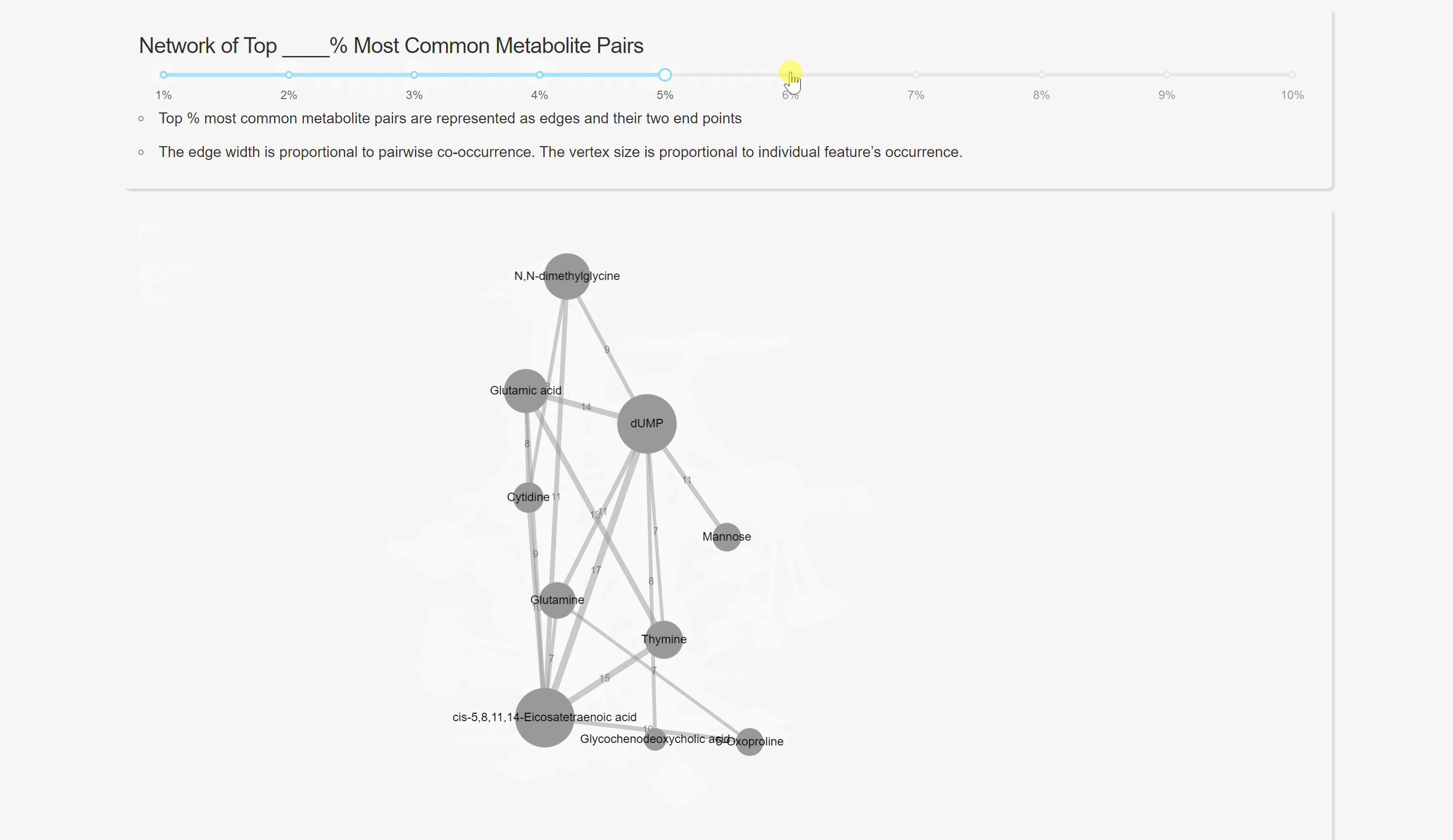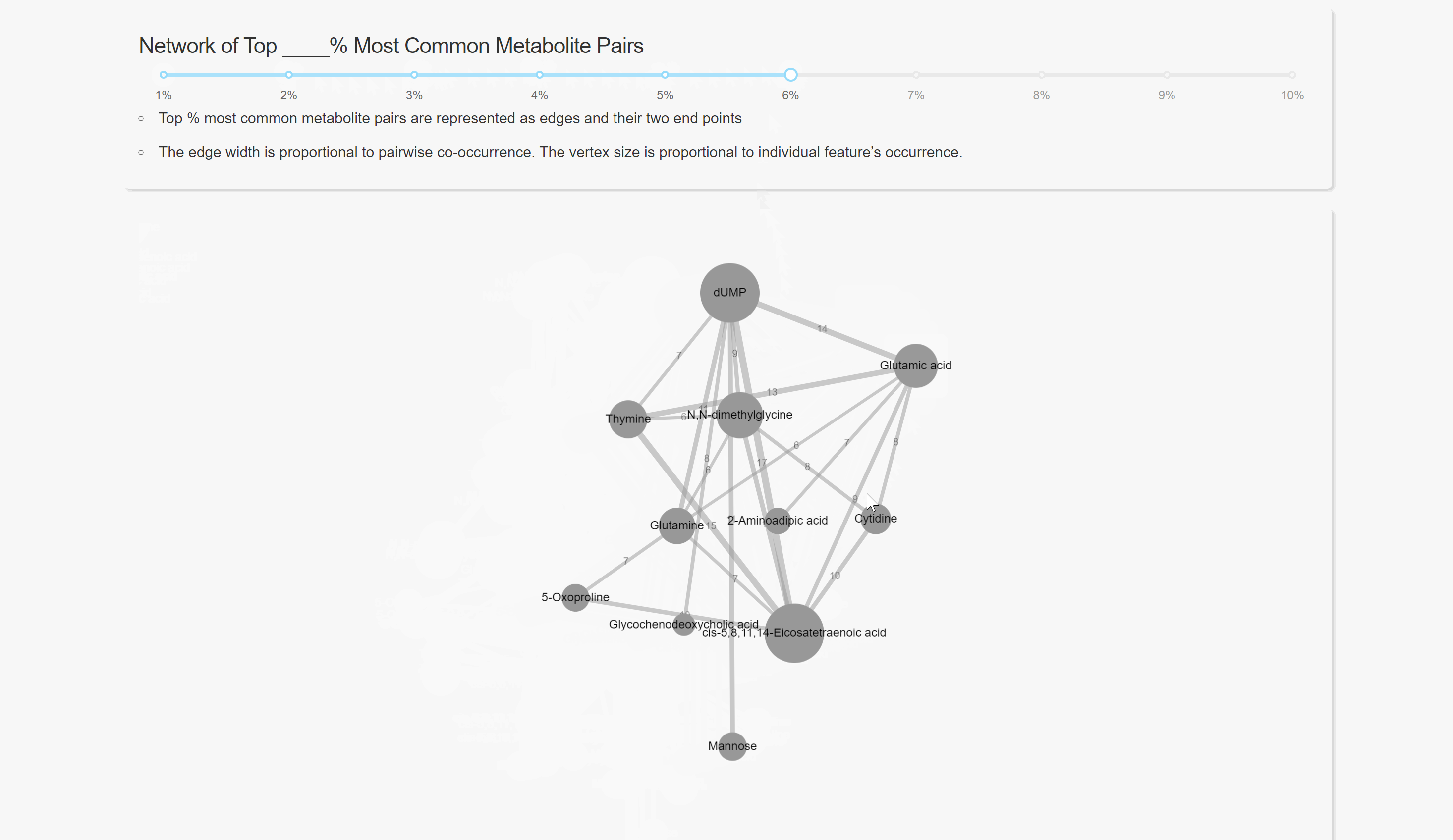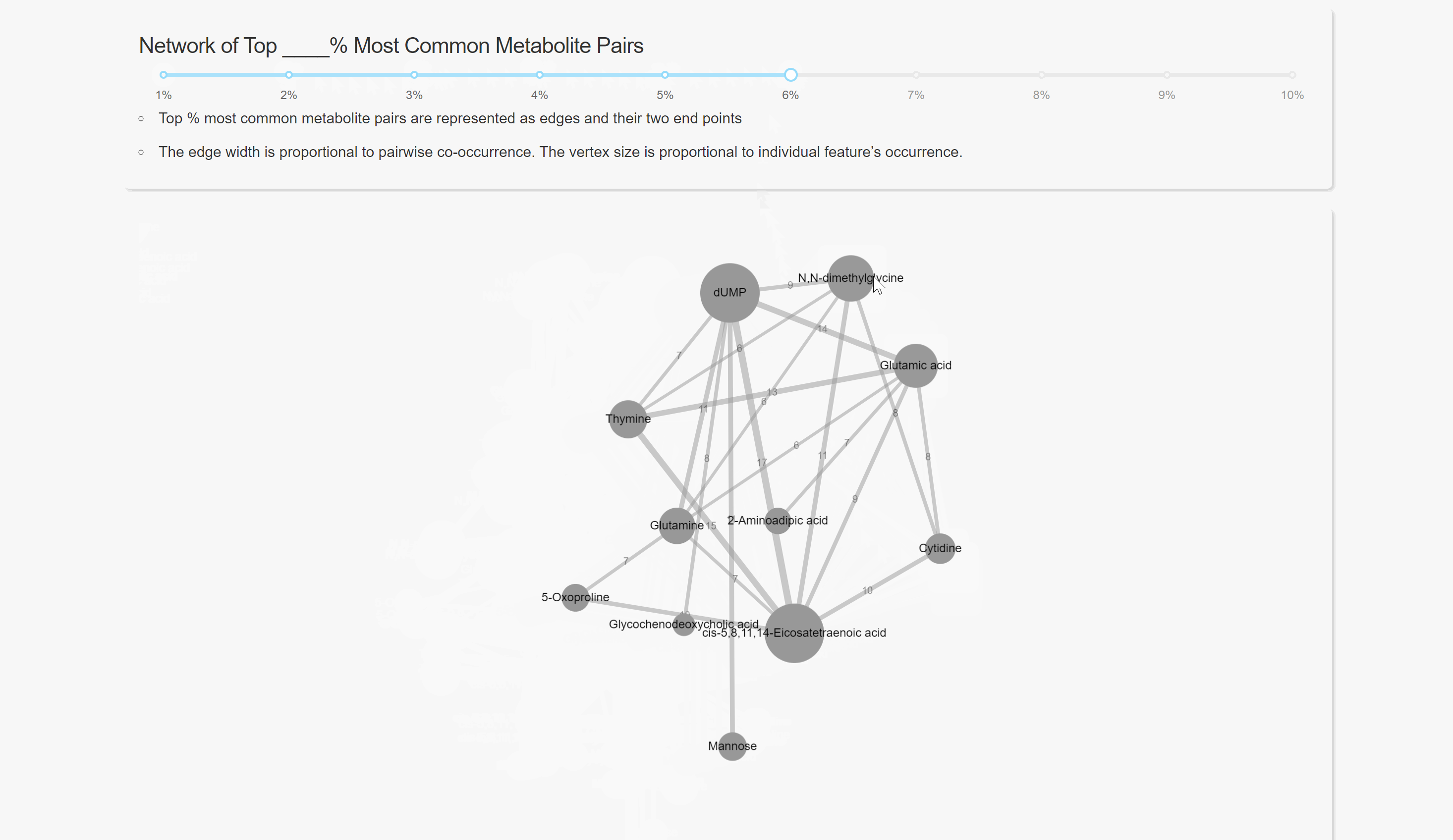
Pairwise Co-occurrence Analysis
Network Analysis
Linear_Genetic_Programming. Authors: Brameier, Markus F., Banzhaf, Wolfgang