FMBIO-Model to-do: explain FMBIO-Model
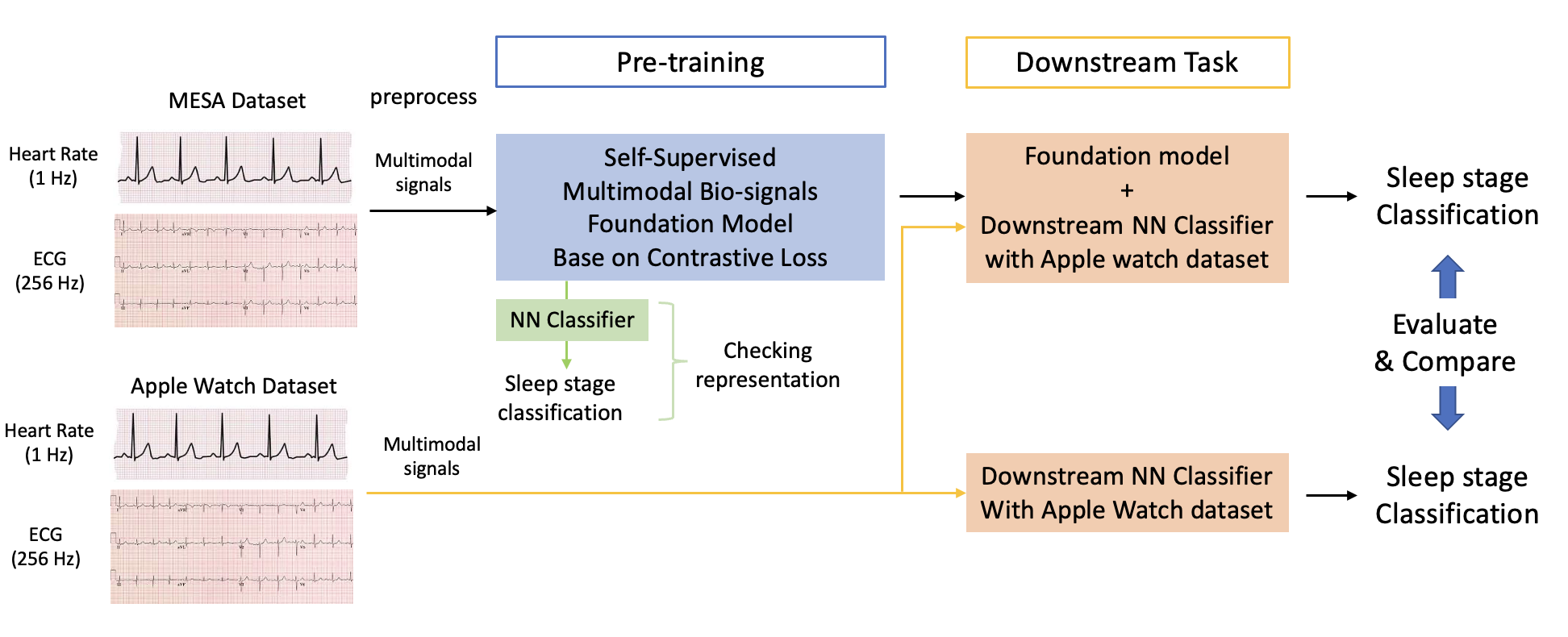
We will attempt to develop the foundation model using bio-signal (ECG, Heart Rate) [1] for applying the sleep stage classification from the personal data of edge devices [2], such as Apple Watch or Fit-bit. It is hard to get high performance by only using personally own data from edge devices and to train the model, as limitation of the amount of data for train and low hardware resources of edge devices. We expect that the foundation model generates informative representative feature from large bio-signal dataset, and it can improve the downstream task in the restricted environment that people cannot share bio-signal data to others.
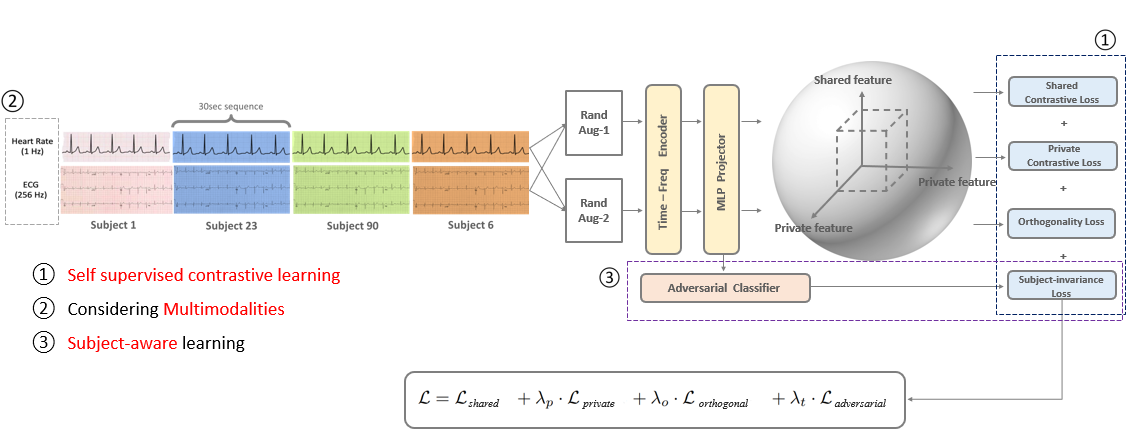
 Our framework has two main strategies:
Our framework has two main strategies:
- MultiModalLoss: develop the foundation model using bio-signal (ECG, Heart Rate)
- Subject-Invariant:
create and activate conda environment named FMBIO with python=3.8.18
conda create -n FMBIO python=3.8 -y
conda activate FMBIO
pip install -r requirements.txtWe used two dataset MESA dataset and Apple Watch dataset. The description of dataset is below.
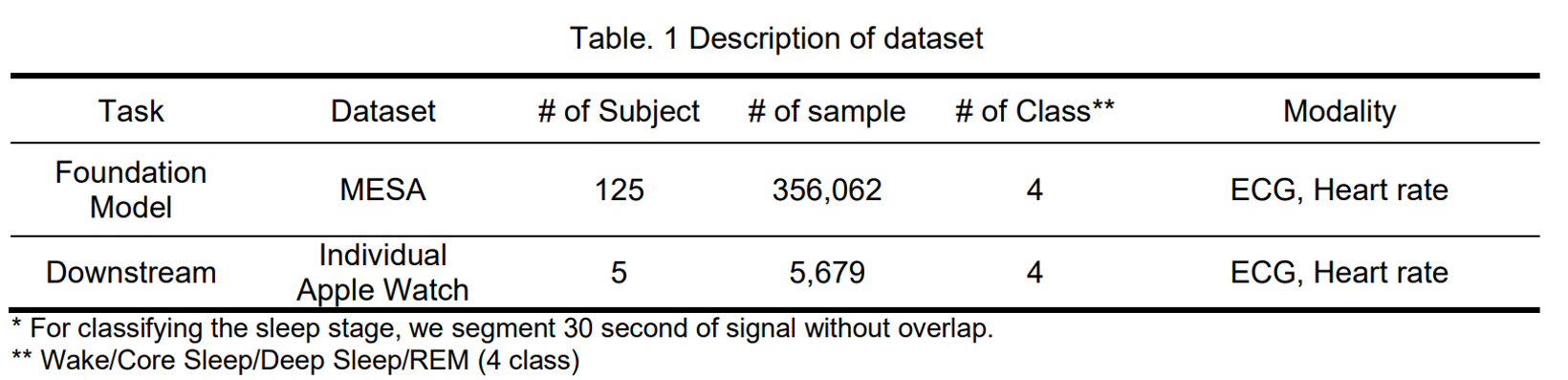 Due to access issue, the MESA dataset MESA Dataset cannot share to others. In addition, we only share 1 subject Apple Watch dataset for demo.
Due to access issue, the MESA dataset MESA Dataset cannot share to others. In addition, we only share 1 subject Apple Watch dataset for demo.
The pre-processing scripts are included in this repo. For preprocessing the MESA dataset and Apple Watch dataset, we cited GitHub
python ./src/foundation/train.pyTrain using Jupyter Notebook basemodel/subj_baseline.ipynb
(1) Evaluation Metrics
| Client | Model | Accuracy | F1-Score |
|---|---|---|---|
| 1 | Base | 0.804 | 0.705 |
| FM | 0.816 | 0.689 | |
| 2 | Base | 0.621 | 0.317 |
| FM | 0.641 | 0.380 | |
| 3 | Base | 0.784 | 0.400 |
| FM | 0.791 | 0.404 | |
| 4 | Base | 0.736 | 0.595 |
| FM | 0.648 | 0.506 | |
| 5 | Base | 0.812 | 0.692 |
| FM | 0.716 | 0.518 |
(2) Confusion Matrix
[1] Large-scale Training of Foundation Models for Wearable Biosignals
[2] Sleep stage prediction with raw acceleration and photoplethysmography heart rate data derived from a consumer wearable device