MUBD-DecoyMaker 3.0: Making Maximal Unbiased Benchmarking Data Sets with Deep Reinforcement Learning
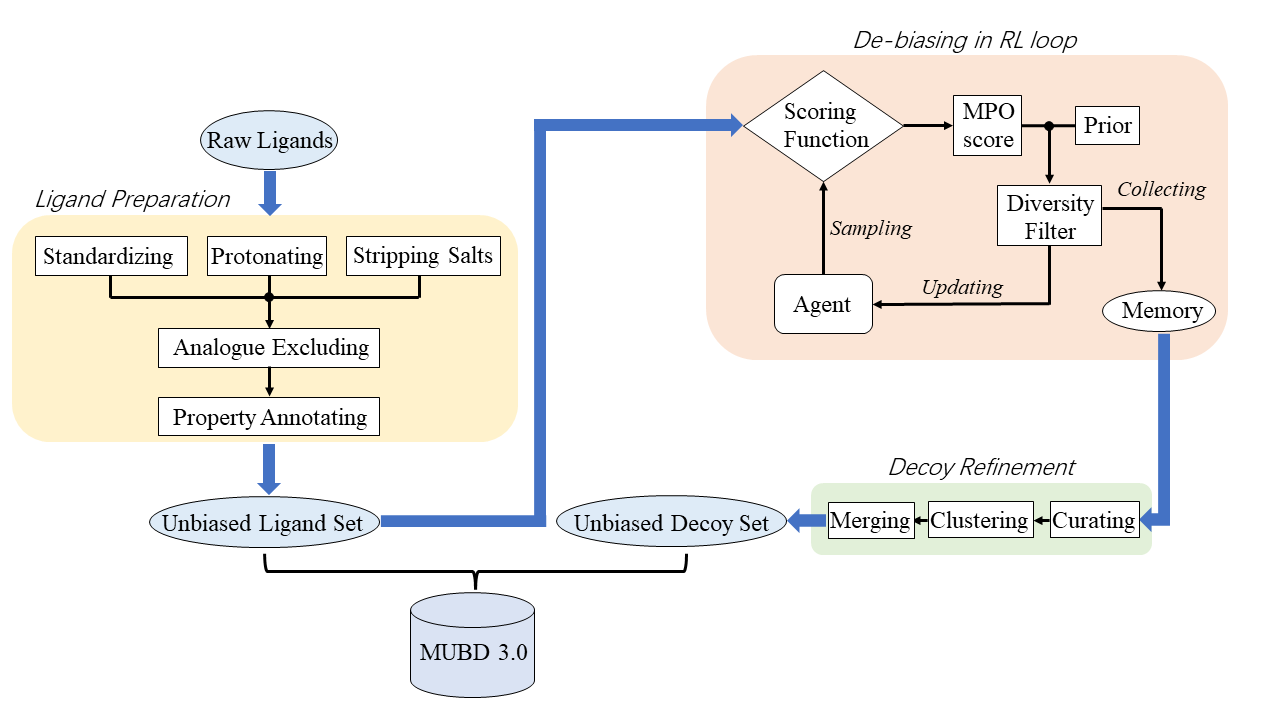
MUBD-DecoyMaker 3.0 is a brand-new computational software to make Maximal Unbiased Benchmarking Data Sets (MUBD) for in silico screening. Compared with our earlier two versions, i.e. MUBD-DECOYMAKER (Pipeline Pilot-based version, or MUBD-DecoyMaker 1.0) and MUBD-DecoyMaker 2.0, MUBD-DecoyMaker 3.0 has two noteworthy features:
-
Virtual molecules generated by recurrent neural netwrok (RNN)-based molecular generator with reinforcement learning (RL), instead of chemical library molecules, constitue the unbiased decoy set (UDS) component of MUBD.
-
The criteria (or rule) for an ideal decoy previously defined in the earlier versions are integrated into a new scoring function for RL to fine-tune the generator.
Below is how to implement and run MUBD-DecoyMaker 3.0.
As REINVENT is used to make virtual decoys of MUBD 3.0, users are required to install this tool as instructed. The corresponding conda environment named reinvent.v3.2 is created for virtual decoy generation. Please note we used and modified the source code from reinvent-chemistry and reinvent-scoring, Copyright 2021 Atanas Patronov, licensed under the Apache 2.0 license, here in order to include our scoring functions specific for MUBD. Another conda environment named MUBD3.0 is also created for preprocessing and postprocessing.
-
Install REINVENT.
-
Clone this repository and navigate to it:
$ git clone https://github.com/Sooooooap/MUBD3.0.git
$ cd MUBD3.0- Replace the packages
reinvent_chemistryandreinvent_scoringwith our modified ones:
$ conda activate reinvent.v3.2
$ pip show reinvent_chemistry # Location: ~/anaconda3/envs/reinvent.v3.2/lib/python3.7/site-packages
$ cp -r reinvent_chemistry/ reinvent_scoring/ ~/anaconda3/envs/reinvent.v3.2/lib/python3.7/site-packages- Create the
condaenvironment calledMUBD3.0:
$ conda env create -f MUBD3.0.ymlACM Agonists is used as a test case to demonstrate how to build MUBD 3.0 with MUBD-DecoyMaker 3.0. All the test files are in the directory of ./resources/.
Run build_uls.py to process the raw ligand set. This script takes the raw ligands in SMILES representation as input (raw_actives.smi) and puts out the unbiased ligand set (Diverse_ligands.csv). Four more files regarding ligand properties, i.e. Diverse_ligands_PS.csv, Diverse_ligands_PS_maxmin.csv, Diverse_ligands_sims_maxmin.txt and Diverse_ligands_len.txt, are also generated and stored in the directory of ./output/ULS/.
IMPORTANT: Ligand curation, including molecule standardization, salt removal and protonization at a specific range of pH (implemented by Dimorphite-DL), is required if the ligands are not curated. For ligand curation, we provide the --curate option for build_uls.py. Please note the raw ligands in this test case are curated. Also, users may use --help option to see all the available options.
$ conda activate MUBD3.0
(MUBD3.0) $ python build_uls.py --i resources/raw_actives.smimk_config.py writes out the configurations for the generation of MUBD3.0 virtual decoys. We provide gen_decoys.sh to iterate over all the ligands and set the configurations specific for each of them. Please make sure the </path/to/REINVENT> in gen_decoys.sh is replaced with the user-defined directory.
$ chmod +x ./gen_decoys.sh
$ conda activate reinvent.v3.2
(reinvent.v3.2) $ ./gen_decoys.shThe file with the directory of ./output/UDS/auto_train/ligand_*/results/scaffold_memory.csv contains the potential decoy set specific for the ligand_*. The potential decoy set is refined by SMILES curation and structural clustering (script: curating_clustering.py). Then the unbiased decoys for each ligand were annotated with the properties and merged (script: merge_decoys.py) to consitute the whole data set (Final_decoys.csv). We provide build_uds.sh to automatically run the above mentioned scripts.
$ chmod +x ./build_uds.sh
$ conda activate MUBD3.0
(MUBD3.0) $ ./build_uds.shThe MUBD 3.0 is validated and measured with four basic metrics. We provide validate.py to perform the validation and store the results in the directory of ./validation/results/:
(MUBD3.0) $ python validate.pyWe appreciate that authors who published REINVENT 2.0: An AI Tool for De Novo Drug Design make REINVENT open to the community. Our work is based on this tool and please consider citing their work if you use MUBD-DecoyMaker 3.0 in your research.
We appreciate that authors who published Dimorphite-DL: an open-source program for enumerating the ionization states of drug-like small molecules make Dimorphite-DL open to the community. We use this tool to protonate the raw actives and please consider citing their work if you use this function.