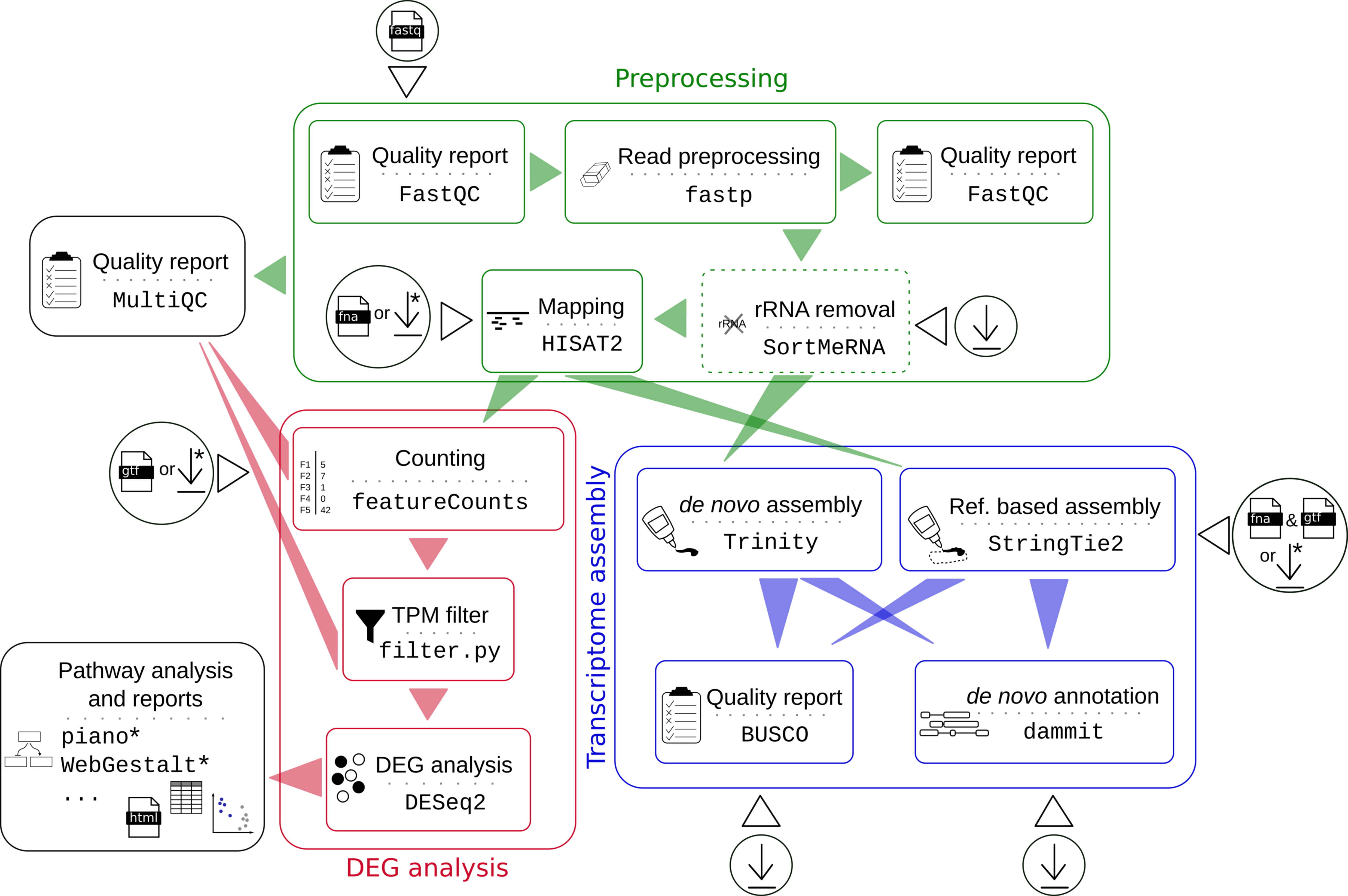
 Figure 1. Workflow. The user can decide after preprocessing to run a differential gene expression (DEG) analysis or a transcriptome assembly. Circles symbolize input data and download icons symbolize automated download of resources. Steps marked by asterisks are currently only available for some species. See here for a list of references for the used tools and please consider to cite them as well.
Figure 1. Workflow. The user can decide after preprocessing to run a differential gene expression (DEG) analysis or a transcriptome assembly. Circles symbolize input data and download icons symbolize automated download of resources. Steps marked by asterisks are currently only available for some species. See here for a list of references for the used tools and please consider to cite them as well.
Table of Contents
- Quick installation
- Quick start
- Usage
- Workflow control
- Profiles/configuration options
- Monitoring
- Output
- Working offline
- Help message
- Known bugs and issues
- [Problems with
SortMeRNA/HISAT2error (#141, #116)](#problems-with-sortmerna-hisat2-error-141-116) - Latency problems on HPCs, issue (#79)
- [Problems with
- Citation
The pipeline is written in Nextflow, which can be used on any POSIX compatible system (Linux, OS X, etc). Windows system is supported through WSL. You need Nextflow installed and either conda, Docker, or Singularity to run the steps of the pipeline:
-
Install
Nextflowclick here for a bash one-liner
wget -qO- https://get.nextflow.io | bash # In the case you don’t have wget # curl -s https://get.nextflow.io | bash
-
Install
condaclick here for a bash two-liner for Miniconda3 Linux 64-bit
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh bash Miniconda3-latest-Linux-x86_64.sh
OR
-
Install
condaclick here for a bash two-liner for Miniconda3 Linux 64-bit
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh bash Miniconda3-latest-Linux-x86_64.sh
-
Install
Nextflowviacondaclick here to see how to do that
conda create -n nextflow -c bioconda nextflow conda active nextflow
For transcriptome assembly you have to install also Docker or Singularity.
-
You can try to simply install
Singularityviacondaas wellclick for an example command
conda create -n singularity -c conda-forge singularity conda active singularity
or if you already have a conda environment for nextflow:
conda activate nextflow conda install -c conda-forge singularity
A system admin-configured Singularity installation should be preferred in comparison to an own local conda installation. Please ask your sys admin!
All other dependencies and tools will be installed within the pipeline via conda, Docker or Singularity depending on the profile you run (see below).
# conda active nextflow
nextflow run hoelzer-lab/rnaflow -profile test,conda,local... performs
- a differential gene expression analysis with sub-sampled human read data,
- on a reduced human genome and annotation (chromosome 1, 10 and 11),
- comparing two conditions (MAQCA, MAQCB),
- with a local execution (uses max. 4 cores in total and 8GB) and
condadependency management.
Resource usage
For a local test run (with 30 cores in total at maximum):
nextflow run hoelzer-lab/rnaflow -profile test,conda,local -w work \
--max_cores 30 --cores 10 --softlink_results -r masterwe observed the following resource usage including downloads and conda environment creation for each process:
- Total runtime
- 25m 56s
- Physical memory (RAM), max.
- 3.015 GB at process
hisat2index - Virtual memory (RAM + Disk swap), max.
- 10.16 GB at process
hisat2
A detailed HTML report automatically produced the pipeline can be found here.
nextflow run hoelzer-lab/rnaflow --helpnextflow pull hoelzer-lab/rnaflowWe recommend to use a stable release of the pipeline:
nextflow pull hoelzer-lab/rnaflow -r <RELEASE>nextflow run hoelzer-lab/rnaflow --reads input.csv --autodownload hsa --pathway hsa --max_cores 6 --cores 2with --autodownload <hsa|mmu|mau|eco> build-in species, or define your own genome reference and annotation files in CSV files:
nextflow run hoelzer-lab/rnaflow --reads input.csv --genome fastas.csv --annotation gtfs.csv --max_cores 6 --cores 2Genomes and annotations from --autodownload, --genome and --annotation are concatenated.
By default, all possible comparisons are performed. Use --deg to change this.
--pathway <hsa|mmu|mau> performs downstream pathway analysis. Available are WebGestalt set enrichment analysis (GSEA) for hsa, piano GSEA with different settings and consensus scoring for hsa, mmu and mau.
Specify your read files in FASTQ format with --reads input.csv. The file input.csv has to look like this for single-end reads (just leave R2 empty):
Sample,R1,R2,Condition,Source,Strandedness
mock_rep1,/path/to/reads/mock1.fastq.gz,,mock,,0
mock_rep2,/path/to/reads/mock2.fastq.gz,,mock,,0
mock_rep3,/path/to/reads/mock3.fastq.gz,,mock,,0
treated_rep1,/path/to/reads/treat1.fastq.gz,,treated,,0
treated_rep2,/path/to/reads/treat2.fastq.gz,,treated,,0
treated_rep3,/path/to/reads/treat3.fastq.gz,,treated,,0
and for paired-end reads, like this:
Sample,R1,R2,Condition,Source,Strandedness
mock_rep1,/path/to/reads/mock1_1.fastq,/path/to/reads/mock1_2.fastq,mock,A,0
mock_rep2,/path/to/reads/mock2_1.fastq,/path/to/reads/mock2_2.fastq,mock,B,0
mock_rep3,/path/to/reads/mock3_1.fastq,/path/to/reads/mock3_2.fastq,mock,C,0
treated_rep1,/path/to/reads/treat1_1.fastq,/path/to/reads/treat1_2.fastq,treated,A,0
treated_rep2,/path/to/reads/treat2_1.fastq,/path/to/reads/treat2_2.fastq,treated,B,0
treated_rep3,/path/to/reads/treat3_1.fastq,/path/to/reads/treat3_2.fastq,treated,C,0
The first line is a required header. Read files can be compressed (.gz). You need at least two replicates for each condition to run the pipeline. Source labels are optional - the header is still required, the value can be empty as in the single-end example above. Source labels can be used to define the corresponding experiment even more precisely for improved differential expression testing, e.g. if RNA-Seq samples come from different Conditions (e.g. tissues) but the same Sources (e.g. patients). Still, the comparison will be performed between the Conditions but the Source information is additionally used in designing the DESeq2 experiment. Source labels also extend the heatmap sample annotation. Strandedness for the samples can optionally be defined directly in the csv or via the commandline parameter --strand. Where the strandedness column can be any value from: 0 = unstranded, 1 = stranded, 2 = reversely stranded, [default: 0]. Note that if strandedness is provided via the input CSV and the commandline parameter, the value from the command line will be used for the run.
If you don't use one of the build-in species, specify your genomes via --genome fastas.csv, with fastas.csv looking like this:
/path/to/reference_genome1.fasta
/path/to/reference_genome2.fasta
and --annotation gtfs.csv with gtfs.csv looking like this:
/path/to/reference_annotation_1.gtf
/path/to/reference_annotation_2.gtf
You can add a build-in species to your defined genomes and annotation with --autodownload xxx.
We provide a small set of build-in species for which the genome and annotation files are automatically downloaded from Ensembl with --autodownload xxx. Please let us know, we can easily add other species.
| Species | three-letter shortcut | Genome | Annotation |
|---|---|---|---|
| Homo sapiens | hsa * |
Homo_sapiens.GRCh38.98 | Homo_sapiens.GRCh38.dna.primary_assembly |
| Mus musculus | mmu * |
Mus_musculus.GRCm38.99 | Mus_musculus.GRCm38.dna.primary_assembly |
| Mesocricetus auratus | mau * |
Mesocricetus_auratus.MesAur1.0.100 | Mesocricetus_auratus.MesAur1.0.dna.toplevel |
| Escherichia coli | eco |
Escherichia_coli_k_12.ASM80076v1.45 | Escherichia_coli_k_12.ASM80076v1.dna.toplevel |
* Downstream pathway analysis availible via --pathway xxx.
To adjust the handling of multiple-mapped reads during the feature counting process you can use:
--featurecounts_additional_params '-t exon -g gene_id -M'
The default handling is to only count uniquely mapped reads via featureCounts. With the above flag set featureCounts will also count multi-mapped reads.
Per default, all possible pairwise comparisons in one direction are performed. Thus, when A is compared against B the pipeline will not automatically compare B vs. A which will anyway only change the direction of the finally resulting fold changes. To change this, please define the needed comparison with --deg comparisons.csv, where each line contains a pairwise comparison:
Condition1,Condition2
conditionX,conditionY
conditionA,conditionB
conditionB,conditionA
The first line is a required header.
You can easily resume your run in case of changes to the parameters or inputs. Nextflow will try to not recalculate steps that are already done:
nextflow run hoelzer-lab/rnaflow -profile test,conda,local -resume
Nextflow will need access to the working directory where temporary calculations are stored. Per default, this is set to work but can be adjusted via -w /path/to/any/workdir. In addition, the .nextflow.log file is needed to resume a run, thus, this will only work if you resume the run from the same folder where you started it.
--skip_sortmerna # skip rRNA removal via SortMeRNA [default false]
--skip_read_preprocessing # skip preprocessing with fastp [default: false]
--fastp_additional_params # additional parameters for fastp [default '-5 -3 -W 4 -M 20 -l 15 -x -n 5 -z 6']
--hisat2_additional_params # additional parameters for HISAT2
--featurecounts_additional_params # additional parameters for FeatureCounts [default: -t gene -g gene_id]--strand # strandness for counting with featureCounts: 0 (unstranded), 1 (stranded) and 2 (reversely stranded) [default 0]
--tpm # threshold for TPM (transcripts per million) filter [default 1]
--deg # a CSV file following the pattern: conditionX,conditionY
--pathway # perform different downstream pathway analysis for the species hsa|mmu|mau
--feature_id_type # ID type for downstream analysis [default: ensembl_gene_id]--assembly # switch to transcriptome assembly
--busco_db # BUSCO database ['euarchontoglires' or path to existing DB]
--dammit_uniref90 # add UniRef90 to dammit databases, takes long [false]
--rna # activate directRNA mode for ONT transcriptome assembly [default: false (cDNA)]Per default, the pipeline is locally executed with conda dependency management (corresponds to -profile local,conda). Adjust this setting by combining an executer option with an engine option, e.g. -profile local,conda or -profile slurm,conda. We also provide container support, see below.
... or how to schedule your workload.
Currently implemented are local, slurm and lsf executions.
You can customize local with this parameters:
--cores # cores for one process [default 1]
--max_cores # max. cores used in total [default allAvailable]
--memory # max. memory in GB for local use [default 8 GB]... or in which environment to run the tools.
Currently implemented are conda, Docker and Singularity. For transcriptome assembly some tools need to be run with Docker or Singularity.
You can switch between different engines via -profile, for example:
nextflow run hoelzer-lab/rnaflow -profile test,local,conda
nextflow run hoelzer-lab/rnaflow -profile test,local,docker
nextflow run hoelzer-lab/rnaflow -profile test,slurm,singularity
As a best practice for a local execution, we recommend to run the pipeline with --cores 1 --max_cores 1 the first time you use Singularity, because we experienced issues when generating the Singularity images in parallel the first time the pipeline is executed with this engine option. It is also possible to run the pipeline once with --setup set. In setup mode all the necessary files (DBs, reference files and images) are being downloaded and set up.
You can customize where conda environments are stored using
--condaCacheDir /path/to/dirand where Singularity images are stored via
--singularityCacheDir /path/to/dirDocker images are stored based on your system configuration.
To monitor your computations the pipeline can be connected to Nextflow Tower. You need an user access token to connect your Tower account with the pipeline. Simply generate a login using your email and then click the link send to this address.
"Nextflow Tower does not require a password or registration procedure. Just provide your email address and we'll send you an authentication link to login. That's all!"
Once logged in, click on your avatar on the top right corner and select "Your tokens". Generate a token or copy the default one and set the environment variable:
export TOWER_ACCESS_TOKEN=<YOUR_COPIED_TOKEN>
export NXF_VER=20.10.0You can save this command to your .bashrc or .profile to not need to enter it again.
Now run:
nextflow run hoelzer-lab/rnaflow -profile test,local,conda -with-towerAlternatively, you can also activate the Tower connection within the nextflow.config file located in the root GitHub directory:
tower {
accessToken = ''
enabled = true
} You can also directly enter your access token here instead of generating the above environment variable.
The result folder is structured by each step and tool (results/step/tool) as follows:
results/
├── 01-Trimming
│ └── fastp trimmed reads
├── 02-rRNARemoval
│ └── SortMeRNA rRNA-free (and trimmed) reads
├── 03-Mapping
│ └── HISAT2 mapping results in BAM format with index files (BAI)
├── 04-Counting
│ └── featureCounts counting table
├── 05-CountingFilter
│ └── TPM counting table with additional TPM value; formatted counting table filtered by TPM
├── 06-Annotation filtered annotation; gene id, name and bio type mapping
├── 07-DifferentialExpression
│ └── DESeq2 see below
├── 08-Assembly
│ └── de_novo
│ └── Trinity Trinity assembly (with --assembly)
├── 09-RNA-Seq_Annotation BUSCO, dammit and StringTie2 results (with --assembly)
├── Logs Nextflow execution timeline and workflow report
└── Summary MultiQC report
Please note, that 08-Assembly and 09-RNA-Seq_Annotation are part of the transcriptome assembly branch (--assembly). Here, steps 04 to 07 are currently not applicable.
The DESeq2 result is structured as follows:
07-DifferentialExpression/
└── DESeq2
├── data
│ ├── counts normalized, transformed counts; size factors table
│ └── input DESeq2 input summary
├── deseq2.Rout R log file
├── MAQCA_vs_MAQCB results for pairwise comparison (here exemplarily for the -profile test data set)
│ ├── downstream_analysis
│ │ ├── piano piano results
│ │ └── WebGestalt WebGestalt results
│ ├── input DESeq2 input summary
│ ├── plots
│ │ ├── heatmaps
│ │ ├── MA
│ │ ├── PCA
│ │ ├── sample2sample
│ │ └── volcano
│ ├── reports DESeq2 result HTML table; summary report
│ └── results raw and filtered DESeq2 result in CSV and XLSX format; DEG analysis summary
└── plots heatmaps and PCA of all samples
We provide DESeq2 normalized, regularized log (rlog), variance stabilized (vsd) and log2(n+1) (ntd) transformed count tables (DESeq2/data/counts).
For each comparison (specified with --deg or, per default, all possible pairwise comparisons in one direction), a new folder X_vs_Y is created. This also describes the direction of the comparison, e.g., the log2FoldChange describes the change of a gene A under condition Y with respect to the gene under condition X. For example, a log2FoldChange of +2 for gene A would tell you that this gene is 2-fold upregulated when we compare condition X vs. condition Y. The gene A is higher expressed in samples belonging to condition X.
Downstream analysis (--pathway xxx) are currently provided for some species: GSEA consensus scoring with piano for Homo sapiens (hsa), Mus musculus (mmu) and Mesocricetus auratus (mau); and WebGestalt GSEA for Homo sapiens and Mus musculus.
In case you don't have an internet connection, here is a workaround to this issue for manual download and copying of external recourses:
- Genomes and annotation can also be specified via
--genomeand--annotation, see here. - For
BUSCOit is a simple download, see here withbusco_db = 'euarchontoglires_odb9'as default. - For
SortMeRNAanddammitthe tools must be installed. Version specifications can be found here and there, the code to create the databases here and there withbusco_db = 'euarchontoglires_odb9'dammit_uniref90 = falseas default. - Downstream analysis with
pianoandWebGestaltcurrently need an internet connection in any case. If no connection is availablepianoandWebGestaltare skipped.
RNAflow looks up the files here:
nextflow-autodownload-databases # default: `permanentCacheDir = 'nextflow-autodownload-databases'`
└── databases
└── busco
└── <busco_db>.tar.gz
└── dammit
└── <busco_db>.tar.gz
└── uniref90 # in case of `dammit_uniref90 = true`
└── <busco_db>.tar.gz
└── sortmerna
└── data
└── rRNA_databases
click here to see the complete help message
Usage examples:
nextflow run hoelzer-lab/rnaflow -profile test,local,conda
nextflow run hoelzer-lab/rnaflow --cores 4 --reads input.csv --autodownload mmu --pathway mmu
nextflow run hoelzer-lab/rnaflow --cores 4 --reads input.csv --autodownload eco --assembly
nextflow run hoelzer-lab/rnaflow --cores 4 --reads input.csv --genome fasta_virus.csv --annotation gtf_virus.csv --autodownload hsa --pathway hsa
Genomes and annotations from --autodownload, --genome and --annotation are concatenated.
Input:
--reads A CSV file following the pattern: Sample,R1,R2,Condition,Source,Strandedness - read mode is detected automatically
(check terminal output if correctly assigned)
Per default, all possible comparisons of conditions in one direction are made. Use --deg to change.
--autodownload Specifies the species identifier for automated download [default: ]
Currently supported are:
- hsa [Ensembl: Homo_sapiens.GRCh38.dna.primary_assembly | Homo_sapiens.GRCh38.98]
- eco [Ensembl: Escherichia_coli_k_12.ASM80076v1.dna.toplevel | Escherichia_coli_k_12.ASM80076v1.45]
- mmu [Ensembl: Mus_musculus.GRCm38.dna.primary_assembly | Mus_musculus.GRCm38.99.gtf]
- mau [Ensembl: Mesocricetus_auratus.MesAur1.0.dna.toplevel | Mesocricetus_auratus.MesAur1.0.100]
--species Specifies the species identifier for downstream path analysis. (DEPRECATED)
If `--include_species` is set, reference genome and annotation are added and automatically downloaded. [default: ]
Currently supported are:
- hsa [Ensembl: Homo_sapiens.GRCh38.dna.primary_assembly | Homo_sapiens.GRCh38.98]
- eco [Ensembl: Escherichia_coli_k_12.ASM80076v1.dna.toplevel | Escherichia_coli_k_12.ASM80076v1.45]
- mmu [Ensembl: Mus_musculus.GRCm38.dna.primary_assembly | Mus_musculus.GRCm38.99.gtf]
- mau [Ensembl: Mesocricetus_auratus.MesAur1.0.dna.toplevel | Mesocricetus_auratus.MesAur1.0.100]
--genome CSV file with genome reference FASTA files (one path in each line)
If set, --annotation must also be set.
--annotation CSV file with genome annotation GTF files (one path in each line)
--include_species Either --species or --genome/--annotation need to be used. Both input seetings can be also combined to use genome and annotation of
supported species in addition to --genome and --annotation [default: false]
Preprocessing options:
--fastp_additional_params additional parameters for fastp [default: -5 -3 -W 4 -M 20 -l 15 -x -n 5 -z 6]
--skip_sortmerna skip rRNA removal via SortMeRNA [default: false]
--skip_read_preprocessing skip preprocessing with fastp [default: false]
--hisat2_additional_params additional parameters for HISAT2 [default: ]
--featurecounts_additional_params additional parameters for FeatureCounts [default: -t gene -g gene_id]
DEG analysis options:
--strand 0 (unstranded), 1 (stranded) and 2 (reversely stranded) [default: 0]
This will overwrite the optional strandedness defined in the input CSV file.
--tpm Threshold for TPM (transcripts per million) filter. A feature is discared, if for all conditions the mean TPM value of all
corresponding samples in this condition is below the threshold. [default: 1]
--deg A CSV file following the pattern: conditionX,conditionY
Each line stands for one differential gene expression comparison.
Must match the 'Condition' labels defined in the CSV file provided via --reads.
--pathway Perform different downstream pathway analysis for the species. [default: ]
Currently supported are:
- hsa | Homo sapiens
- mmu | Mus musculus
- mau | Mesocricetus auratus
--feature_id_type ID type for downstream analysis [default: ensembl_gene_id]
Transcriptome assembly options:
--assembly Perform de novo and reference-based transcriptome assembly instead of DEG analysis [default: false]
--busco_db The database used with BUSCO [default: euarchontoglires_odb9]
Full list of available data sets at https://busco-data.ezlab.org/v5/data/lineages/
--dammit_uniref90 Add UniRef90 to the dammit databases (time consuming!) [default: false]
--rna Activate directRNA mode for ONT transcriptome assembly [default: false (cDNA)]
Computing options:
--cores Max cores per process for local use [default: 1]
--max_cores Max cores used on the machine for local use [default: 4]
--memory Max memory in GB for local use [default: 8 GB]
--output Name of the result folder [default: results]
Caching:
--permanentCacheDir Location for auto-download data like databases [default: nextflow-autodownload-databases]
--condaCacheDir Location for storing the conda environments [default: conda]
--singularityCacheDir Location for storing the singularity images [default: singularity]
--workdir Working directory for all intermediate results [default: null] (DEPRECATED: use `-w your/workdir` instead)
--softlink_results Softlink result files instead of copying.
--setup Download all necessary DB, reference and image files without running the pipeline. [default: false]
Nextflow options:
-with-tower Activate monitoring via Nextflow Tower (needs TOWER_ACCESS_TOKEN set).
-with-report rep.html CPU / RAM usage (may cause errors).
-with-dag chart.html Generates a flowchart for the process tree.
-with-timeline time.html Timeline (may cause errors).
Execution/Engine profiles:
The pipeline supports profiles to run via different Executers and Engines e.g.: -profile local,conda
Executer (choose one):
local
slurm
lsf
latency
Engines (choose one):
conda
mamba
docker
singularity
Per default: -profile local,conda is executed.
For a test run (~ 15 min), add "test" to the profile, e.g. -profile test,local,conda.
The command will create all conda environments and download and run test data.
We also provide some pre-configured profiles for certain HPC environments:
ara (slurm, conda and parameter customization)
The pipeline fails with something like
this
Error executing process > 'preprocess:hisat2 (2)'
Caused by:
Missing output file(s) `22_rep4_summary.log` expected by process `preprocess:hisat2 (2)`
Command executed:
hisat2 -x reference -1 22_rep4.R1.other.fastq.gz -2 22_rep4.R2.other.fastq.gz -p 60 --new-summary --summary-file 22_rep4_summary.log | samtools view -bS | samtools sort -o 22_rep4.sorted.bam -T tmp --threads 60
Command exit status:
0
Command output:
(empty)
Command error:
Error: Read AFFFJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJ has more quality values than read characters.
terminate called after throwing an instance of 'int'
Aborted (core dumped)
(ERR): hisat2-align exited with value 134
[bam_sort_core] merging from 0 files and 60 in-memory blocks...
grep: warning: GREP_OPTIONS is deprecated; please use an alias or script
Work dir:
/tmp/nextflow-work-as11798/2f/4a5b7060530705c2697bdf3eec73a4
Tip: when you have fixed the problem you can continue the execution adding the option `-resume` to the run command line
- Often encountered when running in
screenortmux - Nextflow's
-bgoption does not help
- Skip
SortMeRNAwith--skip_sortmerna - Reads can be cleand beforhand e.g. with CLEAN
Latency problems on HPCs, issue (#79)
Latency related problems with Nextflow might occur when running on HPC systems, where Nextflow expects files to be available before they are fully written to the file system. In these cases Nextflow might get stuck or report missing output or input files to some processes:
ERROR ~ Error executing process > 'some_process'
Caused by:
Missing output file(s) `some_process.out` expected by process `some_process`
- Often encountered when running on HPC systems
Please try running the pipeline with the latency profile activated, just add it to the profiles you already defined:
-profile slurm,conda,latency
If you use RNAflow please cite:
Marie Lataretu and Martin Hölzer. "RNAflow: An effective and simple RNA-Seq differential gene expression pipeline using Nextflow". Genes 2020, 11(12), 1487; https://doi.org/10.3390/genes11121487