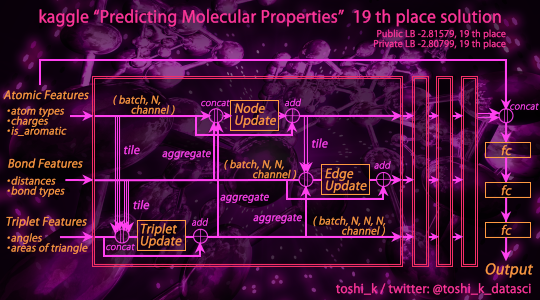
My solution in this Kaggle competition "Predicting Molecular Properties", 19th place.
I used xyz2mol to parse molecular structure.
source/01_preprocess/xyz2mol.py is forked from below repository.
I employed jo's tips to handle xyz2mol in source/01_preprocess/xyz2mol_jo.py
- Using RDKit for Atomic Feature and Visualization
https://www.kaggle.com/sunhwan/using-rdkit-for-atomic-feature-and-visualization
I used train_ob_charges.csv and test_ob_charges.csv which are output of Alexandre's notebook. Please put them in the input directory when you run my code.
- V7 Estimation of Mulliken Charges with Open Babel
https://www.kaggle.com/asauve/v7-estimation-of-mulliken-charges-with-open-babel
Even though, my solution doesn't depend on chainer-chemistry directly, my implementations are inspired by it.
- Chainer Chemistry: A Library for Deep Learning in Biology and Chemistry
https://github.com/pfnet-research/chainer-chemistry
- Neural Message Passing with Edge Updates for Predicting Properties of Molecules and Materials
Peter Bjørn Jørgensen, Karsten Wedel Jacobsen, Mikkel N. Schmidt
https://arxiv.org/abs/1806.03146 - Weisfeiler and Leman Go Neural: Higher-order Graph Neural Networks
Christopher Morris, Martin Ritzert, Matthias Fey, William L. Hamilton, Jan Eric Lenssen, Gaurav Rattan, Martin Grohe
https://arxiv.org/abs/1810.02244