- Introduction
- Installation
- How CURE works
- Quick usage examples
- Output files
- Estimating trees from output files
- Citation
- License
CURE is an automated and parallel pipeline for the Curation of UltraconseRved Elements (UCEs) for species-tree reconstruction. It is an automation/adaptation of the strategies proposed by Van Dam et al. 2021 (named GeneRegion strategy), and Freitas et al. 2021 (name UCERegion strategy).
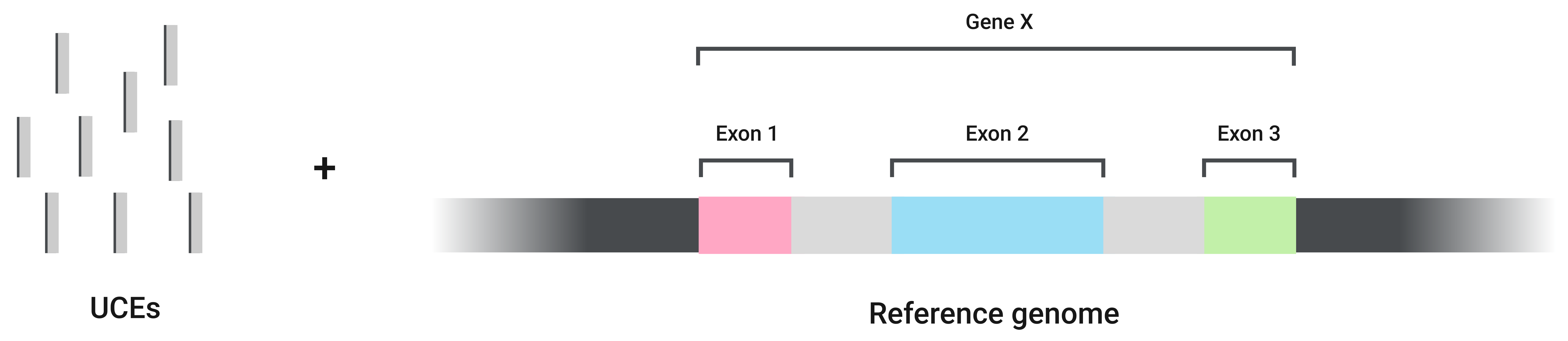
In the GeneRegion strategy (Van Dam et al. 2021), CURE performs the curing process based on the genes in which each UCE is located. CURE can do it in two different ways:
- by gene: concatenates all UCEs from the same gene and treats different genic regions (exons and introns) as different partitions;
- by genic region: concatenates all UCEs from the same exons or introns of the same gene.
When using the GeneRegion strategy, the default behavior of CURE is to run these two ways but this can be changed. The input files for the GeneRegion pipeline are the baits file used for UCE sequencing, the reference genome, an annotation file, and the UCE alignments produced by phyluce.
In the UCERegion strategy (Freitas et al. 2021), CURE performs the curing process based on the internal regions of each UCE (right flank, core, and left flank). It runs SWSC-EN (Tagliacollo & Lanfear 2018) in parallel to speed up the process and creates charsets considering the left flank, core, and right flank as different partitions for each locus in the dataset.
We recommend the installation of CURE with conda for the automatic installation of all dependencies. First, clone this repo in your local machine and enter the created directory:
git clone https://github.com/vhfsantos/CURE.git
cd CURE
Then, create a conda environment for CURE using the cure.yml, and ensure all scripts have execution permission:
conda env create -n cure --file misc/cure.yml
chmod +x CURE
chmod +x scripts/*
After done all installations, activate the CURE environment and run CURE with no arguments. CURE will tell you if any dependencies are missing.
conda activate cure
./CURE
*You should always include the full path to the executable file
The main inputs for this strategy are the UCE alignments and an annotated reference genome (note that CURE also needs to be provided with the baits file used for the UCE sequencing)
The first step of CURE is running a custom version of the uce_type tool described by Van Dam et al. 2021 and available at the Cal Academy's repository.
Note:
uce_typeis distributed by us with CURE. No previous installation of this tools is required.
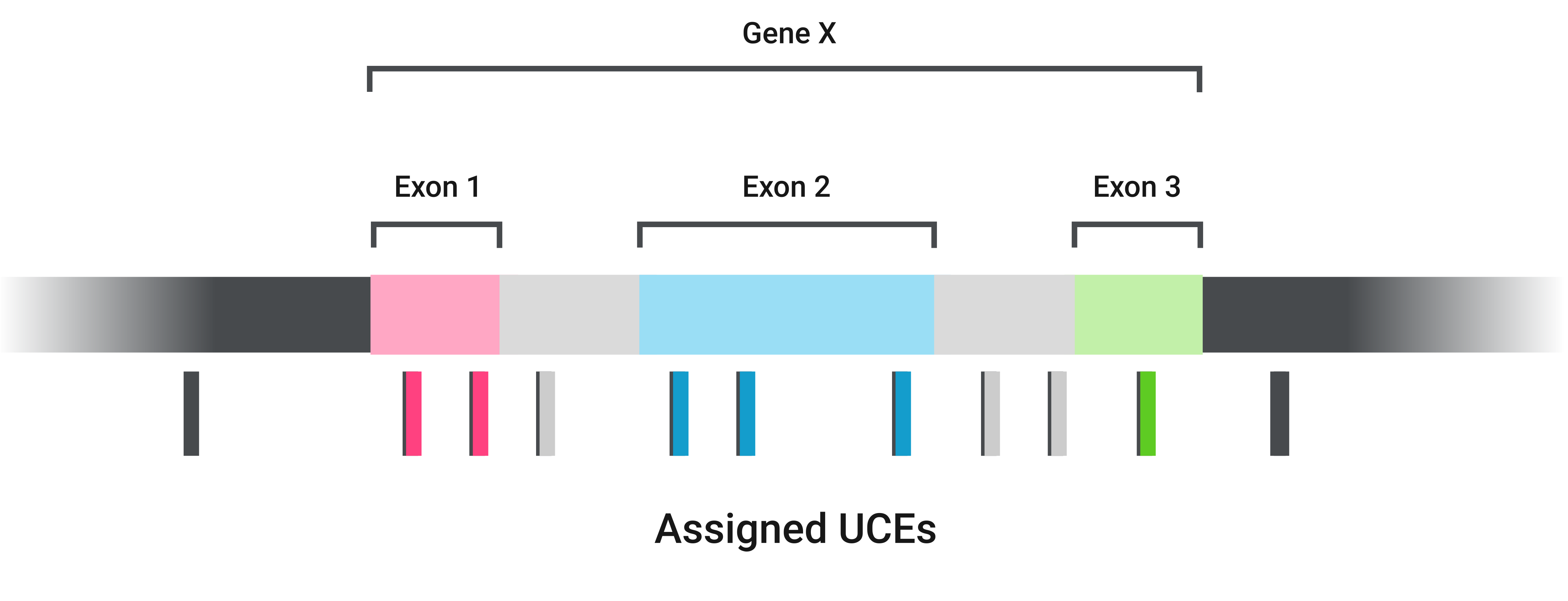
Briefly, this step assigns each UCE to an exon, intron, or intergenic region of the given reference genome
Then CURE parses the results and merges the UCEs in two different ways: by gene and by region.
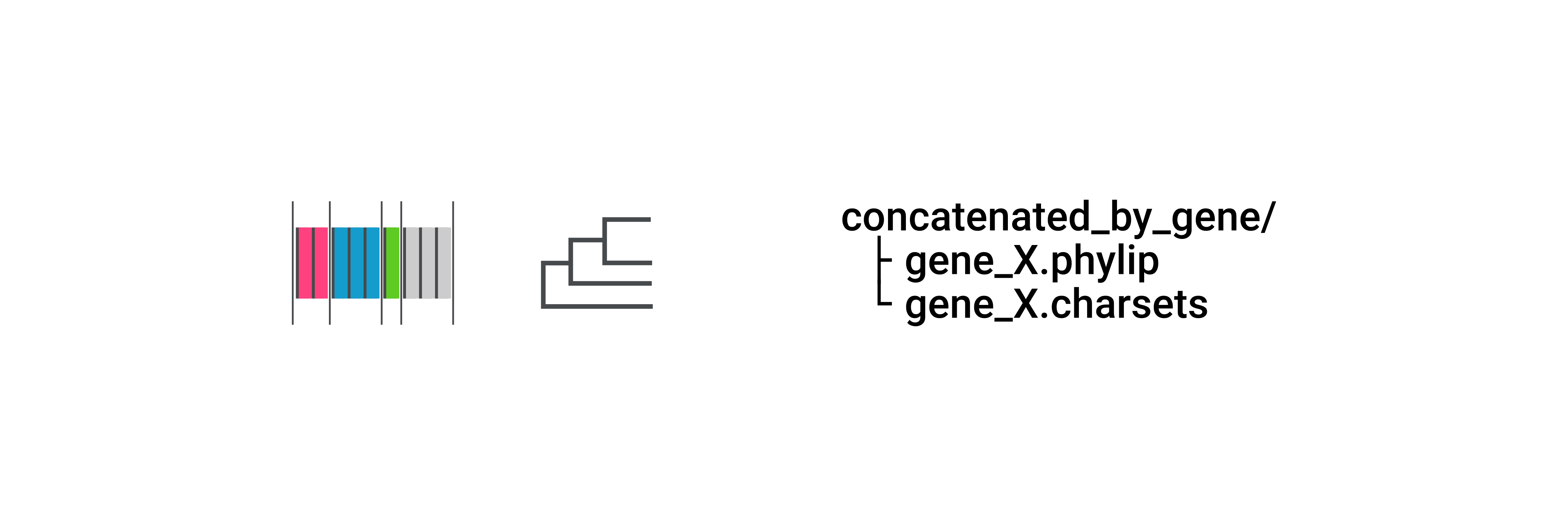
When concatenating by gene, CURE merges all UCEs from the same gene and treats different regions (exons and introns) as different partitions (Note that different introns are placed under the same partition). It stores the results in phylip format inside the concatenated_by_gene/ directory.
Further phylogenetic analysis of UCEs merged with this approach would yield a phylogenetic tree for each gene.
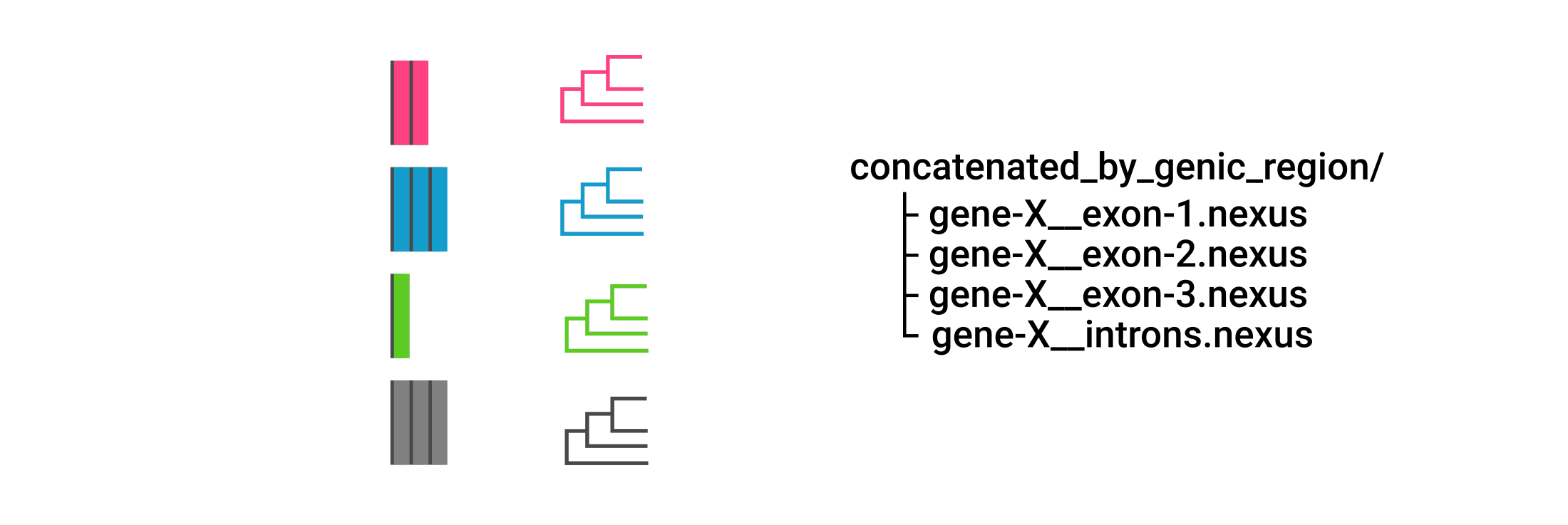
When concatenating by genic region, CURE merges only UCEs from the region of the same gene.
It stores the results in nexus format inside the concatenated_by_genic_region/ directory.
Further phylogenetic analysis of UCEs merged with this approach would yield several phylogenetic trees, one originating from each region.
For any of the two concatenating approaches, CURE leaves unmerged UCEs in intergenic regions.
These UCEs are just copied to the intergenic_regions/ directory.
For this strategy, you need to provide a folder with all the individual alignments you want to use, in nexus format (could be all your alignments or a subset).
CURE runs SWSC-EN in parallel and use PHYLUCE to split the alignments according to regions identified by SWSC.
Then, CURE re-concatenates them, creating a charset file to be used in phylogenetic analyses to generate your gene trees (see Estimating trees from output files), treating each UCE region (left flank, core, and right flank) as different partitions.
Note:
SWSCis distributed by us with CURE. No previous installation of this tools is required.
You can test CURE with the test dataset. It usually takes ~1 minute to run with 2 threads. With the command line below, CURE will run the GeneRegion strategy, concatenating both by gene and by genic region.
./CURE GeneRegion --baits test_data/baits.fasta \
--reference test_data/ref.fa \
--gff test_data/ref.gff \
--phyluce-nexus test_data/uce_nexus/ \
--output ./CURE-GeneRegion-outputBy default, CURE runs the GeneRegion strategy with both concatenating approaches( by gene and by genic region).
However, you can raise the --only-by-gene or --only-by-genic-region flag to select only a single approach
./CURE GeneRegion --baits test_data/baits.fasta \
--reference test_data/ref.fa \
--gff test_data/ref.gff \
--phyluce-nexus test_data/uce_nexus/ \
--output ./CURE-GeneRegion-output \
--only-by-gene./CURE GeneRegion --baits test_data/baits.fasta \
--reference test_data/ref.fa \
--gff test_data/ref.gff \
--phyluce-nexus test_data/uce_nexus/ \
--output ./CURE-GeneRegion-output \
--only-by-genic-regionTo run the test dataset for the UCERegion strategy, use the following command line:
./CURE UCERegion --phyluce-nexus test_data/uce_nexus/ \
--output ./CURE-UCERegion-outputThis takes ~10 minutes with 6 threads.
The main output files produced by CURE are the alignments of concatenated and cured UCEs. Each of the two strategies, however, produces different output files.
CURE UCERegion generates three output subdirectories:
logfiles/: stores all the log files generated in the analysispartitioned-uces/: stores all the alignments and their respective charsets filesconcatenated-uces/: stores the concatenated alignment, a charset file with the results of SWSC-EN in nexus format and a input file for a putative downstream analysis with PartitionFinder2
If you run the GeneRegion strategy without --only-by-gene or --only-by-genic-region, both of the concatenating approaches will be done.
In this case, your output-dir will contain concatenated-by-gene/ and concatenated-by-genic-region/ dirs.
If you raised any of these flags, only the corresponding dir will be created.
Besides, the GeneRegion strategy creates the intergenic-regions/ dir containing unmerged UCEs assigned to intergenic regions.
Alignments in concatenated-by-region/ and intergenic-regions/ dir are in NEXUS format.
Alignments in concatenated-by-gene/ are in PHYLIP format, and its charsets are in NEXUS format.
To avoid troubles with further phylogenetic analysis, CURE replaces "-" with "_" in the gene and exon ID.
Secondary outputs of this strategy include CURE-exons.txt, CURE-introns.txt, and CURE-intergenic.txt, which contains the names of UCEs assigned to each genic region, as well as the region ID (for exons) and gene ID (for exons and introns).
The CURE-intergenic.txt file contains only the UCE names.
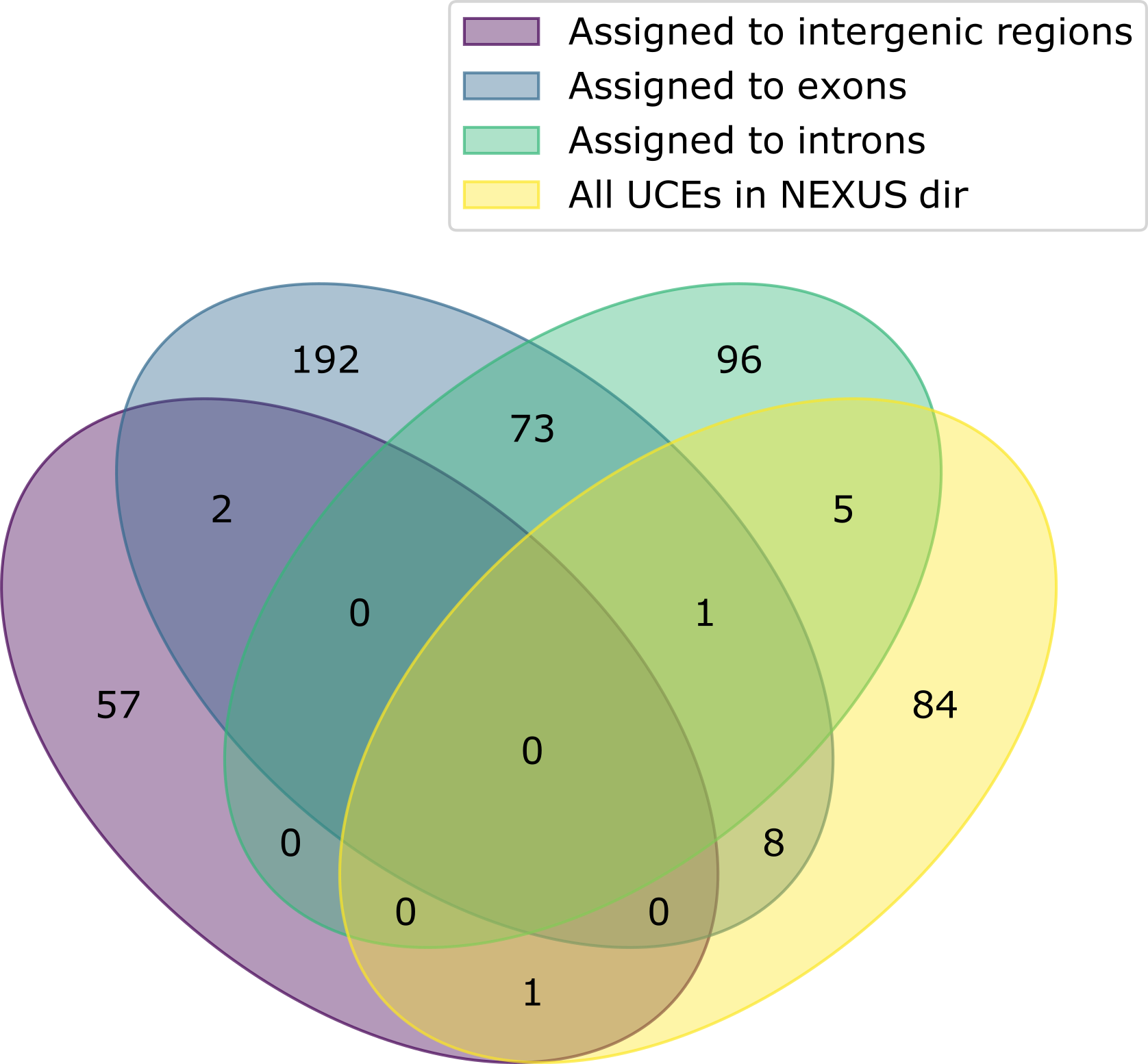
CURE outputs the CURE_stats.csv and CURE_stats.pdf files summarizing the total number of UCEs assigned to each region.
This information is stored in a table-like format in the .csv file and depicted in a Venn diagram in the .pdf file.
The Venn diagram summarizing the test data looks like this:
The group named "All UCEs in NEXUS dir" represents thee UCEs in the directory supplied with the --phyluce-nexus argument.
Numbers outside this yellow ellipse represent the UCEs present in the baits file that were not present in the --phyluce-nexus directory (probably because they were not recovered upstream, by PHYLUCE)
In practical terms, CURE uses only these UCEs for the curation process.
Note that CURE accounts for exons the UCEs assigned to both exon and intron (1, in this test data). Also, it accounts for intergenic regions the unassigned UCEs (84, in this test data).
CURE also maintains in the output directory the files produces by the uce_kit pipeline (uce_kit_output/ dir)
CURE provides the wrapper script estimate-trees.sh for the estimation of gene trees from the output alignments with IQ-tree, and further summary analysis with ASTRAL.
This script runs IQ-tree in parallel using GNU Parallel following the structure of the CURE output-dir.
Then it prepares all inputs needed for a summary analysis with ASTRAL.
For instance, if you run CURE setting CURE-GeneRegion-output as the output directory for the GeneRegion strategy, and CURE-UCERegion-output for the UCERegion strategy, you can call estimate-trees.sh as the following:
scripts/estimate-trees.sh \
--gene-region-out CURE-GeneRegion-output \
--uce-region-out CURE-UCERegion-output \
--estimated-trees estimated-treesIf you run the GeneRegion strategy with --only-by-gene or --only-by-genic-region flags, you can raise it here as well:
scripts/estimate-trees.sh \
--gene-region-out CURE-GeneRegion-output \
--uce-region-out CURE-UCERegion-output \
--estimated-trees estimated-trees \
--only-by-geneor
scripts/estimate-trees.sh \
--gene-region-out CURE-GeneRegion-output \
--uce-region-out CURE-UCERegion-output \
--estimated-trees estimated-trees \
--only-by-genic-regionNote that you don't need to use both
--gene-region-outand--uce-region-out. If you run CURE with only one of the strategies, you only need to use the appropriate parameter.
Moreover, estimate-trees.sh can be used to estimate trees from alignments from any other source; not necessarily those produced by CURE.
In this case, you only need to use the parameter --custom-alignments instead of --gene-region-out or --uce-region-out.
So if you have a set of alignments (in Phylip, Fasta, or Nexus format) in a directory called input-alignments, and want to run IQ-tree on them, you can call estimate-trees.sh as the following:
scripts/estimate-trees.sh \
--custom-alignments input-alignments/ \
--estimated-trees estimated-treesIf you use CURE in your research, please cite: