SVDSS is a novel method for discovery of structural variants in accurate long reads (e.g PacBio HiFi) using sample-specific strings (SFS).
SFS are the shortest substrings that are unique to one genome, called target, w.r.t another genome, called reference. Here our method utilizes SFS for coarse-grained identification (anchoring) of potential SV sites and performs local partial-order-assembly (POA) of clusters of SFS from such sites to produce accurate SV predictions. We refer to our manuscript on SFS for more details regarding the concept of SFS.
You can "install" SVDSS in two different ways:
- compiling it (use this method if you want SVDSS to be fully optimized)
- downloading a static binary
- installing from conda
To compile and use SVDSS, you need:
- a C++14-compliant compiler (GCC 8.2 or newer)
- make, automake, autoconf
- cmake (>=3.14)
- git
- some other development libraries: zlib, bz2, lzma
- samtools and bcftools (>=1.9)
To install these dependencies:
# On a deb-based system (tested on ubuntu 20.04 and debian 11):
sudo apt install build-essential autoconf cmake git zlib1g-dev libbz2-dev liblzma-dev samtools bcftools
# On a rpm-based system (tested on fedora 35):
sudo dnf install gcc gcc-c++ make automake autoconf cmake git libstdc++-static zlib-devel bzip2-devel xz-devel samtools bcftoolsThe following libraries are needed to build and run SVDSS but they are automatically downloaded and compiled while compiling SVDSS:
- htslib built with libdeflate for BAM processing.
- ksw2 for FASTA and FASTQ processing.
- ropebwt2 for FMD index creation and querying.
- abPOA for POA computation.
- parasail for local alignment of POA consensus.
- rapidfuzz for string similarity computation.
- interval-tree for variant overlap detection and clustering.
To download and install SVDSS (should take ~10 minutes):
git clone https://github.com/Parsoa/SVDSS.git
cd SVDSS
mkdir build ; cd build
cmake -DCMAKE_BUILD_TYPE=Release ..
makeThis will create the SVDSS binary in the root of the repo.
For user convenience, we also provide a static binary for x86_64 linux systems (see Releases) - use at your own risk. If it does not work, please let us know or build it yourself :)
SVDSS is available on bioconda:
conda create -n svdss -c conda-forge -c bioconda svdssThis will create the environment svdss that includes SVDSS and its runtime dependencies (i.e., samtools and bcftools).
Please refer to or use Snakefile/run-svdss.sh.
Index reference/sample:
SVDSS index --fastq/--fasta /path/to/genome/file --index /path/to/output/index/file
Optional arguments:
-b, --binary output index in binary format. Allows for another index to be appended to this index later.
-a, --append /path/to/binary/index append to existing binary index.
Extract SFS from BAM/FASTQ/FASTA files:
SVDSS search --index /path/to/index --fastq/--bam /path/to/input --workdir /output/directory
Optional arguments:
--assemble automatically runs SVDSS assemble on output
Assemble SFS into superstrings:
SVDSS assemble --workdir /path/to/.sfs/files --batches /number/of/SFS/batches
Reconstruct sample:
SVDSS smooth --workdir /output/file/direcotry --bam /path/to/input/bam/file --reference /path/to/reference/genome/fasta
Call SVs:
SVDSS call --workdir /path/to/assembled/.sfs/files --bam /path/to/input/bam/file --reference /path/to/reference/genome/fasta
Optional arguments:
--clipped calls SVs from clipped SFS.
--min-cluster-weight minimum number of supporting superstrings for a call to be reported.
--min-sv-length minimum length of reported SVs. Default is 25. Values < 25 are ignored.
General options:
--threads sets number of threads, default 4.
--version print version information.
--help print this help message.
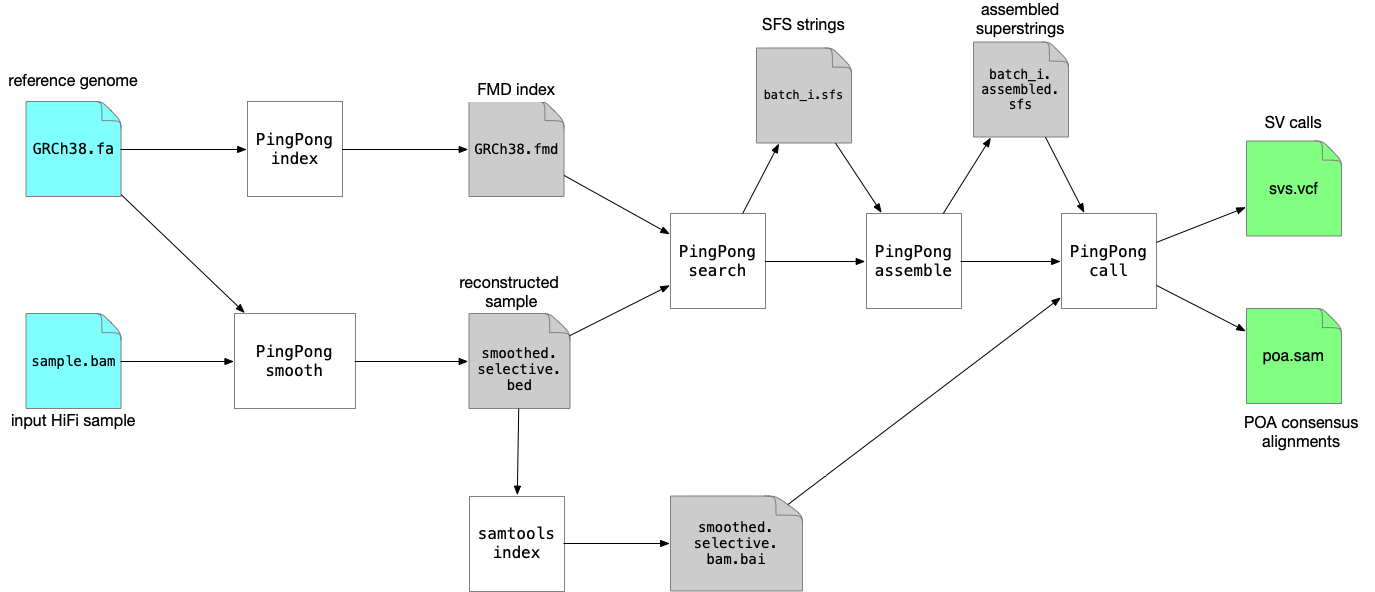
SVDSS requires as input the BAM file of the sample to be genotyped, a reference genome in FASTA format (please use an appropriate reference genome, i.e., if you are not interested in ALT contigs, filter them out or use a reference genome that does not include them). To genotype a sample we need to perform the following steps:
- Build FMD index of reference genome (
SVDSS index) - Smooth the input BAM file (
SVDSS smooth) - Extract SFS from smoothed BAM file (
SVDSS search) - Assemble SFS into superstrings (
SVDSS assemble) - Genotype SVs from the assembled superstrings (
SVDSS call)
In the guide below we assume we are using the reference genome file GRCh38.fa and the input BAM file sample.bam. We assume both files are present in the working directory. All of SVDSS steps must be run in the same directory so we always pass --workdir $PWD for every command.
Note that you can reuse the index from step 1 for any number of samples genotyped against the same reference genome.
Figure below shows the full pipeline of commands that needs to be run:
We will now explain each step in more detail:
Build the FMD index of the reference genome:
SVDSS index --fastq GRCh38.fa --index GRCh38.bwt
The --index option specifies the output file name.
Smoothing removes nearly all SNPs, small indels and sequencing errors from reads. This results in smaller number of SFS being extracted and increases the relevance of extracted SFS to SV discovery significantly. To smooth the sample run:
SVDSS smooth --bam sample.bam --workdir $PWD --reference GRCh38.fa --threads 16
This produces a file named smoothed.selective.bam. This file is sorted in the same order as the input file, however it needs to be indexed again with samtools index. The command also produces two files smoothed_reads.txt and ignored_reads.txt in workdir that contains the ids of reads that were smoothed and ids of reads that didn't have any large (> 20bp) indels in their alignemnts. This information is used by the next step.
To extract SFS run:
SVDSS search --index GRCh38.bwt --bam smoothed.selective.bam --workdir $PWD
This step produces a number of solution_batch_<i>.sfs files. These files include the coordinates of SFS relative to the reads they were extracted from.
To reduce redundancy, overlapping SFS on each reads are merged. Simply run:
SVDSS assemble --workdir $PWD --batches N
Here N is the number of files produces by the previous step. Each .sfs file will be processed independently and output as a solution_batch_<i>.assembled.sfs file.
You can combine SFS extraction and assembly by passing --assemble to SVDSS search. This will automatically run the assembler.
We are now ready to call SVs. Run (note that the input .bam must be sorted and indexed using samtools before running this):
SVDSS call --reference GRCh38.fasta --bam smoothed.selective.bam --workdir $PWD --batches N
You can filter the reported SVs by passing the --min-sv-length and --min-cluster-weight options. These options control the minimum length and minimum number of supporting superstrings for the reported SVs. Higher values for --min-cluster-weight will increase precision at the cost of reducing recall. For a diploid 30x coverage sample, --min-cluster-weight 2 produced the best results in our experiments. For a haploid 30x sample, instead, --min-cluster-weight 4 produced the best results.
This commands output two files: svs_poa.vcf that includes the SV calls and poa.sam which includes alignments of POA contigs to the reference genome (these POA consensus are used to call SVs).
For user convenience, we distribute a Snakefile to run the entire pipeline, from reference + aligned reads to SVs:
# update config.yaml to suit your needs
# run:
snakemake [-n] -j 4
Note: to run this example, samtools and bcftools must be in your path. Running SVDSS on the example data, once downloaded, should take less than 5 minutes.
# Download example data from zenodo
wget https://zenodo.org/record/6563662/files/svdss-data.tar.gz
mkdir -p input
tar xvfz svdss-data.tar.gz -C input
# Download SVDSS binary
wget https://github.com/Parsoa/SVDSS/releases/download/v1.0.3/SVDSS_linux_x86-64
chmod +x SVDSS_linux_x86-64
# Download snakemake workflow and run it
wget https://raw.githubusercontent.com/Parsoa/SVDSS/master/config.yaml
wget https://raw.githubusercontent.com/Parsoa/SVDSS/master/Snakefile
snakemake -p -j 2
# Alternatively, you can use the bash helper script
wget https://raw.githubusercontent.com/Parsoa/SVDSS/master/tests/run-svdss.sh
bash run-svdss.sh ./SVDSS_linux_x86-64 input/22.fa input/22.bam svdss-outputSVDSS was developed by Luca Denti and Parsoa Khorsand.
For inquiries on this software please open an issue or contact either Parsoa Khorsand or Luca Denti.
SVDSS is currently pending peer review. A pre-print is available on BioRxiv.
Instructions on how to reproduce the experiments described in the manuscript can be found here (also provided as submodule of this repository).