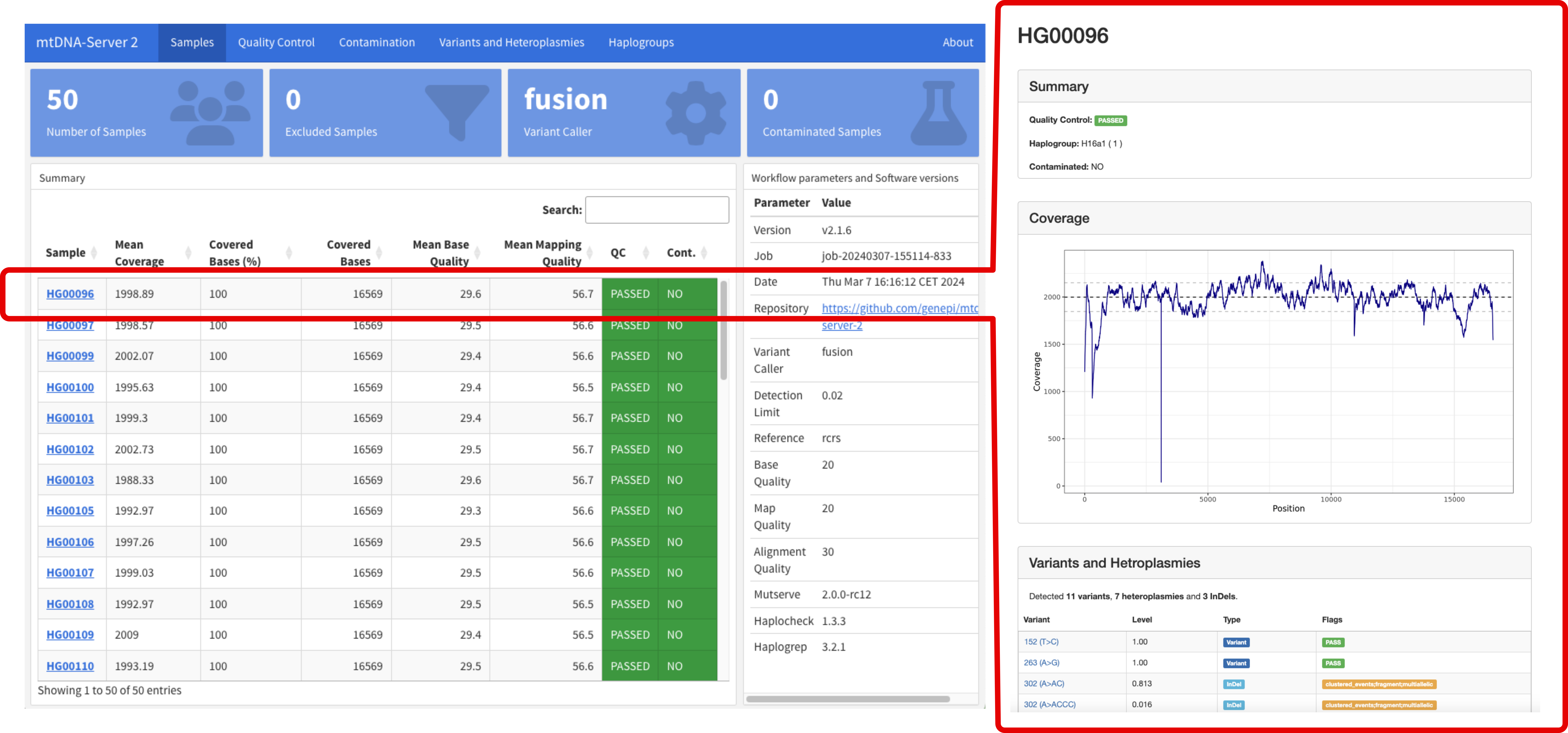
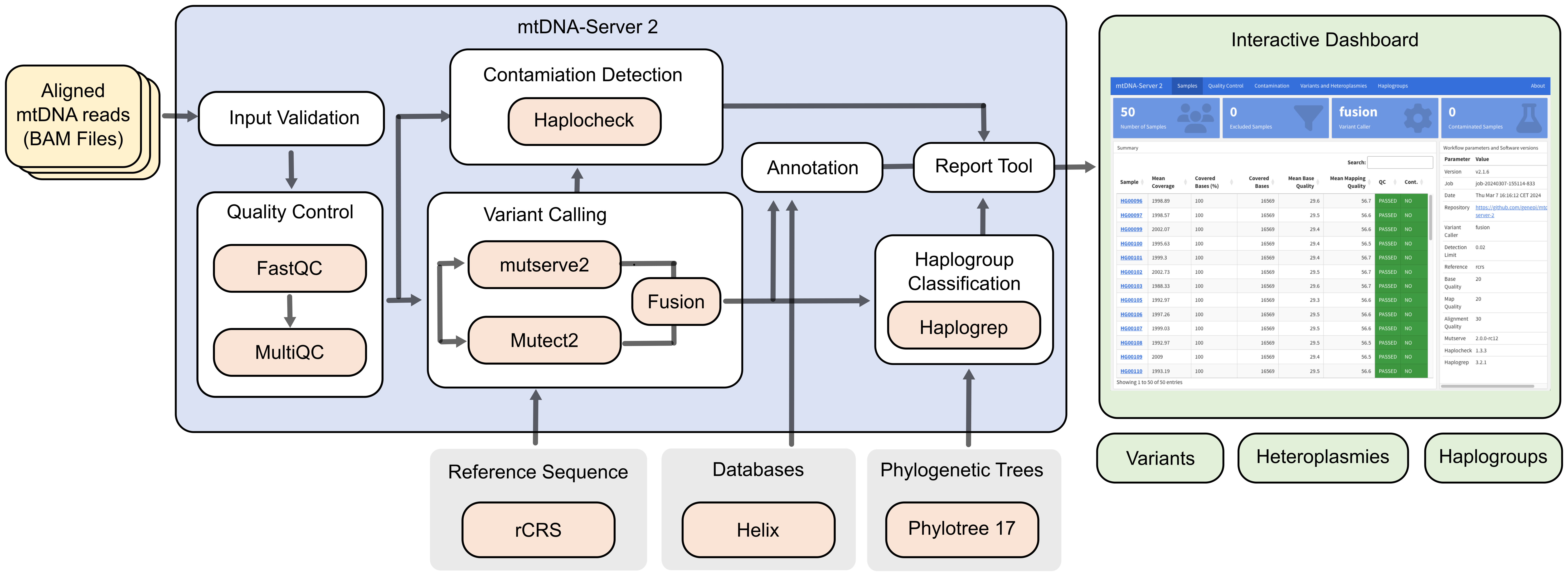
mtDNA-Server 2 is based on Nextflow with the overall goal to accurately detect heteroplasmic and homoplasmic variants in mitochondrial (mtDNA) genomes.
It supports different variant callers and is able to call insertions, deletions (INDELs) and single nucleotide variants (SNVs). Furthmore it includes several modules for (a) quality control and input validation (b) haplogroup classification and contamination detection, (c) minimal VAF estimation based on the coverage to minimize false positives and (d) an interactive analytics dashboard.
mtDNA-Server 2 is hosted as a free service on our mitoverse platform.
Documentation can be accessed here.
- Docker or Singularity
- Java
- Nextflow
nextflow run genepi/mtdna-server-2 -r v2.1.7 -profile test,docker
Weissensteiner H*, Forer L*, Fuchsberger C, Schöpf B, Kloss-Brandstätter A, Specht G, Kronenberg F, Schönherr S: mtDNA-Server: next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic Acids Res. 44:W64-9, 2016. PMID: 27084948
This software was developed at the Institute of Genetic Epidemiology, Medical University of Innsbruck
Sebastian Schoenherr (@seppinho)
Hansi Weissensteiner (@whansi)
MIT Licensed.