-
Notifications
You must be signed in to change notification settings - Fork 4
Home
ProtTrace is a simulation based approach to assess for a protein, the seed, over what evolutionary distances its orthologs can be found by means of sharing a significant sequence similarity. ProtTrace determines for the seed protein a traceability index (TI) that decreases as a function of evolutionary distance. The TI is defined on the interval [0,1], where a TI of 1 indicates that an ortholog should be readily identifiable, whereas a TI of 0 indicates that an ortholog has likely diverged beyond recognition. Once the TI for a protein in a given species is known, it helps to differentiate between the true absence of an ortholog, and its non-detection due to a limited search sensitivity. In the latter case, an increase of the search sensitivity, e.g. by using pHMM based search methods, on the cost of a reduced specificity, can help to identify highly diverged orthologs.
Once the user has specified a seed protein whose traceability should be assessed, a standard ProtTrace analysis includes the following steps
- Compilation of an orthologous group for the seed protein
- Standard: Querying existing ortholog collections (we use OMA groups as a default)
- Expert: Compiling a custom ortholog collection, e.g. by running a targeted ortholog search using HaMStR OneSeq
- Search for Pfam domains in the seed protein's sequence. This analysis requires a local installation of the Pfam database, and of the HMMER package. Note, his information is later used for inferring position-specific constraints on the evolutionary process.
- Inference of the seed proteins' evolutionary parameters. Here, ProtTrace uses first IQ-TREE to compute both the pair-wise Maximum Likelihood distances between the orthologs in the training data with, and a maximum likelihood sequence tree. Using this information, the protTrace then determins the following paramter:
- Rate of insertions and deletions
- length distribution of insertions and deletions
- Substitution rate scaling factor κ
- Simulation of the seed protein evolution with REvolver and determining the traceability curve
- Evolutionary sequence change is simulated in steps of 0.1 substitutions per site up to a total distance of 7.5 substitutions per site.
- Subsequent to each simulation step, the simulated sequence is used as a query in a BlastP search against the entire gene set of the seed species. This step serves to assess whether there is still a significant local sequence similarity between the seed sequence and its evolved instance. The search is a success if the seed protein is among the top five hits.
- Repeat the steps a) and b) 100 times to achieve for each distance the fraction of successes.
- Compute the traceability curve
- In an optional step, the traceability results can be depicted on a phylogenetic tree.
The information flow through the program is shown in Figure 1. A high resolution PDF of the image is available HERE.
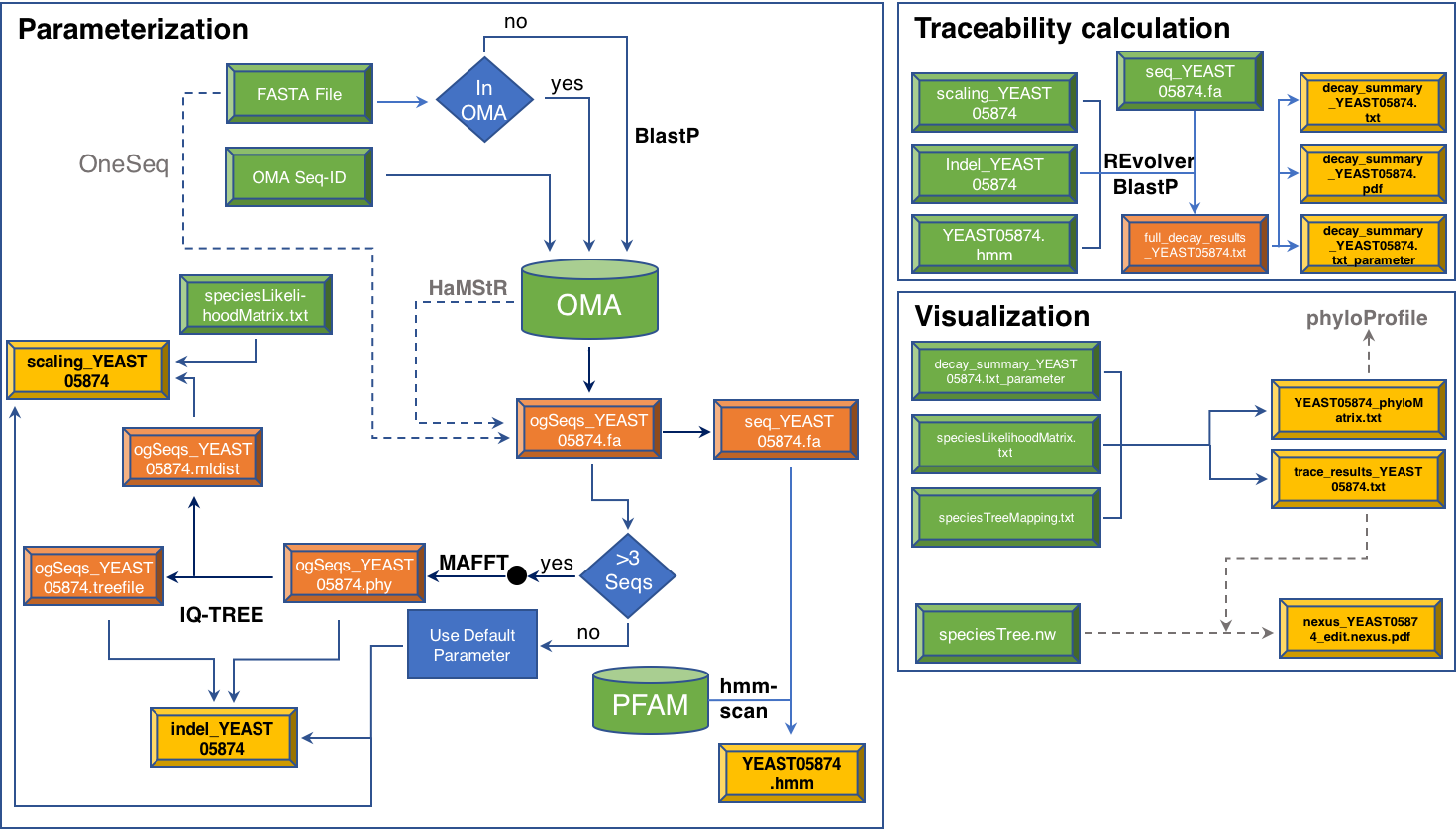
Figure 1 - Workflow of the protTrace analysis The workflow is distinguished into the categories Parameterization, Traceability calculation, and visualization. Boxes in green denote input files, boxes in orange represent meta-information, which is generated in the course of the analysis, and yellow boxes indicate output files that are generated as a result of an analysis. Arrows represent individual analysis steps, where the arrow style indicates whether the analysis step is obligatory (solid), or optional (dashed). Analysis steps that require the calling of external programs are indicated by the program name next to the corresponding arrow. Obligatory dependencies on 3d party software are represented by bold face black program names, those that are optional are indicated by grey font color.
Click HERE to obtain further information about the various output files.
ProtTrace comes along with the requirements for some Accessory Software that needs to be installed along with the ProtTrace package. Note, this is, in almost all cases, standard software for evolutionary sequence analysis, and we trust that most of this software is installed and executable on your system anyway. If not, please find below a detailed instruction of what software is needed and how to install it. Most of the software is available via the BIOCONDA channel of the Conda package manager, so installation on Linux and on MacOS is straightforward.
ProtTrace is written in Java and runs platform-independent. However, some accessory software is limited to Linux / MacOS, such that we recommend installing ProtTrace on either Linux or MacOS. If you are running on Windows, we suggest to install our protTrace Virtual Machine.
The ProtTrace package contains scripts written in different languages. In order to run ProtTrace you need the following resources:
- Python v2.7.13 or higher. Note, ProtTrace will not run under Python 3
- Install also the https://www.dendropy.org/ DendroPy module (can be done via Conda.
- Perl v5 or higher including the following modules
- Getopt::Long
- List::Util
- LWP::Simple
- Java v1.7 or higher
- R v3 or higher
| Program name | Version | Description | Mandatory | BioConda |
|---|---|---|---|---|
| MAFFT | v6 or higher | Multiple Sequence alignment | yes | yes |
| NCBI Blast | v2.7 or higher | Sequence similarity based search | yes | yes |
| HMMER | 3.2 or higher | Sequence similarity based search using Hidden Markov Mode | yes | yes |
| IQTREE | 1.6.7.1 or higher | Phylogenetic tree reconstruction | yes | yes |
| HaMStR OneSeq | v1 or higher | targeted ortholog search | no | no |
We provide two Virtual Machines running Ubuntu Linux that have protTrace and all dependencies installed. See the protTrace-VirtualMachine page for further details.
If you opt for a standard installation of protTrace on your system instead of using the Virtual Machine please follow the guidelines below.
Before installing protTrace, prepare the environment by installing the necessary software dependencies. Click this LINK for a detailed instruction of how to set up the environment using the Conda package management system. Click HERE for a more concise guide.
Use the following steps to create a standard instance of protTrace on your computer. Note, the standard installation works only with pre-existing ortholog assignments and does not use the HaMStR package.
- Install all protTrace dependencies
- Change to a directory where you want the ProtTrace package to be installed
- clone the git repository by typing
git clone https://github.com/BIONF/protTrace.git
This installs all the programs in the directory from which you issued the command.
Change to the protTrace directory and run the script create_conf.pl. This will test for the existence of all software dependencies, and optionally can download the necessary data from the OMA web pages and from Pfam
To see all options for the create_config.pl script, run
perl ./bin/create_config.pl -h
If you run protTrace for the first time, we suggest to run the full set up script by issuing
perl ./bin/create_config.pl -name=prog.config -getPfam -getOma
The script will perform the following steps:
- Check for the software dependencies. If you installed a software in a non-standard path, you can enter the corresponding path interactively.
- Update all paths in the config file prog.config
- offers you the option to set run parameters interactively
- print out a config file that controls the protTrace run. You will find this config file in the same dirctory you started the script create_conf.pl from, and it will have the name you provided in the comman call using the option -name.
- download orthologous group assignment from the OMA database together with the corresponding sequences (option -getOma). In addition, the Pfam-A database will be downloaded (option -getPfam). The data will be placed into the directory protTrace/used_files, and the corresponding paths in the config file.
If all tests succeeded, protTrace will be ready to run. In case, a problem occurs, it will be printed to the screen. On top of this, a log file will be generated.
protTrace requires, by default, orthologous groups assigned by [https://omabrowser.org OMA] together with the protein sequences in Fasta format, and the Pfam database.
- If you have this information already available at you computer, specify the corresponding paths in the protTrace configuration file, either by manually editing the config file, or by running the configuration script create_conf.pl. Make sure that the protein sequences are formatted such that each sequence extends only over one line. See HERE for details about downloading and preparing the OMA ortholog assignments.
- If you are unsure whether or not these files exist on your computer, use the configuration script create_conf.pl provided in the bin directory for downloading and reformatting the files. Use the options
-getPfam -getOma
for this purpose. Note: The script will attempt to download about 6Gb of data, so this may take a while. The files will be placed in the used_files directory.
For running protTrace, you will have to configure the individual run parameters that are then passed on to the program via the config file. For creating an initial config file, run the script
create_config.pl
which is provided in the bin directory of the protTrace distribution. For modifying an existing config file, simply provide the name of the existing file and add the option -update.
create_conf.pl -name=YourConfigFile -update
The script will then guide you through the update procedure.
- In brief, there are seven main parameter classes controlling different steps of the analysis:
- [0] - General Options
- [1] - Preprocessing Settings
- [2] - Advanced Preprocessing Settings
- [3] - Scaling factors
- [4] - Indel parameter
- [5] - Traceability calculation
- [6] - Program paths
- [7] - Paths to files
- You can select one to several of the main classes, by
- providing the corresponding numbers, each separated by a comma
- providing a range, e.g. 1-7 will select all classesOnce the main classes have been selected, the script will then ask you to select the parameter(s) you want to update. Note: If you selected more than one main parameter class, the script will ask you first for each class which parameters you want to set.
- As a last step, the script will ask you to enter your values for the selected parameters. For each parameter it provides you with the current setting, and the default value (if existent), or a brief description of what to enter.
- Once all parameters have been set, the config file will be saved and is ready to use.
See the following code as an example
Once you have completed all installation steps and did run the configuration script create_conf.pl you should be ready to go. We have provided two example files in the directory toy_example with which you can test your protTrace installation.
The most convenient way of starting a protTrace analysis is to provide the program an OMA sequence id. The file test.id contains the OMA id YEAST05874. To start a traceability analysis with this sequence, run protTrace as following:
- check in your protTrace config file that the parameter species is set to YEAST . For the next step, we assume that a config file prot.config is located in the directory toy_example.
- change into the directory toy_example and run protTrace by issuing the following command
../bin/protTrace.py -i test.id -c prot.config
Click HERE to access a summary of the command line output during the protTrace run. The table below summarizes the main information that you should find in your output directory upon a successful completion of the protTrace run.
Optionally, you can start protTrace using a protein sequence in FASTA format as the seed. The file test.fa contains the protein sequence of the human protein ZNT3. protTrace will then, as its first step, use a BLAST search to identify the corresponding OMA identifier for this sequence. To start a traceability analysis with this sequence, run protTrace as following:
- check in your protTrace config file that the parameter species is set to HUMAN. For the next step, we assume that a config file prot2.config is located in the directory toy_example.
- change into the directory toy_example and run protTrace by issuing the following command
../bin/protTrace.py -f test.fasta -c prot2.config
Click HERE to access a summary of the command line output during the protTrace run. The output produced is analogous to the one shown in the table below.
The followign table provides information about the main output files of the protTrace run using YEAST05874 as the seed protein. Meta-results, such as Blast libraries, and protein set collections for the YEAST, which are generated in the course of the analysis are not listed. The links open up the result files as PDF.
| Task | Filename | Description |
|---|---|---|
| Pfam domain annotation | YEAST05874.hmm | File containing the hmmscan output for the seed sequence |
| Ortholog identification | ogIds_YEAST05874.txt | Members of the OMA ortholog group the seed protein belongs to |
| ogSeqs_YEAST05874.fa | Amino acid sequences for the OMA ortholog group | |
| MSA of orthologous sequences | ogSeqs_YEAST05874.phy | MAFFT-linsi alignment of the orthologous sequences |
| Phylogeny reconstruction | ogSeqs_YEAST05874.phy.iqtree | ML tree reconstruction of the orthologous sequence |
| Pairwise distance computation | ogSeqs_YEAST05874.phy.mldist | Computation of the pairwise ML distances between the orthologs |
| Scaling factor | scale_YEAST05874 | Compare pairwise distances between sequences to pairwise distances of species |
| Indel rate | indel_YEAST05874 | Rate and shape parameter of the geometric indel distribution |
| Decay analysis | full_decay_results_YEAST05874.txt | Results of the simulation procedure |
| decay_summary_YEAST05874.txt | Summary of decay analysis over 100 repetitions | |
| decay_summary_YEAST05874.txt.pdf | Graphical display of the traceability curve | |
| trace_results_YEAST05874.txt | This file contains traceabilities for every reference taxon listed in your Xref_mapping_file (species_tree_maping.txt). The third column in the file gives the traceabilities values | |
| Visualization | nexus_YEAST05874_edit.nexus.pdf | Display of the traceability results on the taxonomy tree. The traceability index (TI) of the seed protein in the respective species is color coded from green, representing high a high TI, to yellow, representing intermediate TI, and finally to red, representing a low TI |
| YEAST05874_phyloMatrix.txt | Tabular output of the traceability analysis for upload and visualization in PhyloProfile |